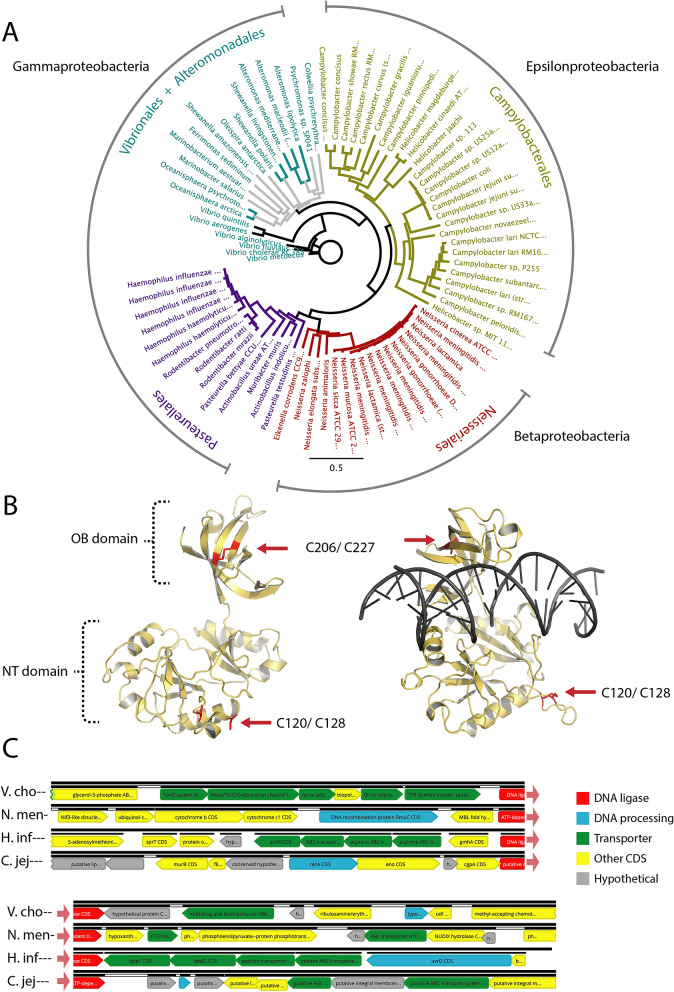
Figure 2.
(A) Phylogenetic analysis of bacterial Lig E sequences coloured by Order. Solid-coloured branches indicate bootstrap values greater than 50% consensus support. Protein identifiers for ligases are given in Supplementary Table 1. (B) Lig E from Campylobacter jejuni modeled in open DNA-free (left) and closed DNA-bound (right) conformations. Potential disulfide bonds in the OB and NT domains are indicated in red. (C) Genomic context of Lig E from pathogenic representatives of different bacterial Orders. Species names are abbreviated as: V. cho, Vibrio cholera: N. men, Neisseria meningitidis; H. infl, Haemophilus influenzae; C. jej, Campylobacter jejuni. Genes are coloured according to the function annotated in the genomic sequence as DNA processing (blue), transporter (green), other known function unrelated to transport or DNA (yellow) or unknown function (grey). The DNA ligase is coloured red for orientation.