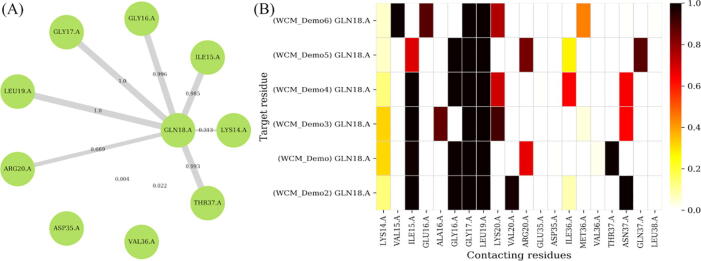
Fig. 4.
Estimating contact frequencies around a single residue in a single and in multiple protein structures, using HIV protease mutants as example. The locus of interest is displayed at the center in panel (A), and is surrounded by its neighbouring residues. Additionally, the edge thicknesses and labels depict the residue contact frequencies obtained from the MD simulations. In both panels, residues are depicted by the three-letter residue code followed by the residue position, a dot and the chain label. In panel (B), multiple related contacts (gathered from the contact mapping tool) are stacked on top of each other along the y-axis, with their neighbours spanning the x-axis.