Summary
Clinical validity assessments of gene-disease associations underpin analysis and reporting in diagnostic genomics, and yet wide variability exists in practice, particularly in use of these assessments for virtual gene panel design and maintenance. Harmonization efforts are hampered by the lack of agreed terminology, agreed gene curation standards, and platforms that can be used to identify and resolve discrepancies at scale. We undertook a systematic comparison of the content of 80 virtual gene panels used in two healthcare systems by multiple diagnostic providers in the United Kingdom and Australia. The process was enabled by a shared curation platform, PanelApp, and resulted in the identification and review of 2,144 discordant gene ratings, demonstrating the utility of sharing structured gene-disease validity assessments and collaborative discordance resolution in establishing national and international consensus.
Clinical validity assessments of gene-disease associations underpin analysis and reporting in diagnostic genomics, and yet wide variability exists in practice, particularly in use of these assessments for virtual gene panel design and maintenance. Harmonization efforts are hampered by the lack of agreed terminology, agreed gene curation standards, and platforms that can be used to identify and resolve discrepancies at scale. We undertook a systematic comparison of the content of 80 virtual gene panels used in two healthcare systems by multiple diagnostic providers in the United Kingdom and Australia. The process was enabled by a shared curation platform, PanelApp, and resulted in the identification and review of 2,144 discordant gene ratings, demonstrating the utility of sharing structured gene-disease validity assessments and collaborative discordance resolution in establishing national and international consensus.
Introduction
Robust gene-disease validity assessments are the foundation of accurate variant interpretation in diagnostic genomics. Reporting variants in genes lacking clinical validity poses risks to patient care through the potential for misinterpretation and erroneous diagnosis. Conversely, the rapid growth in new gene-disease relationships resulting from the deployment of genomic sequencing in research and clinical care presents the risk of missed diagnoses if new knowledge is not continuously integrated into diagnostic systems. Gene knowledge databases commonly used in diagnostic systems have traditionally been curated for other primary purposes such as cataloguing Mendelian gene-disease associations (Orphanet, OMIM) or facilitating diagnosis in specific phenotypic groups (Gene2Phenotype).1 The development of the ClinGen framework significantly advanced evidence-based approaches to gene-disease curation by proposing a scheme for objectively scoring genetic and experimental data.2 The application of this framework by expert groups has resulted in the curation of over 900 gene-disease pairs to date,3, 4, 5, 6, 7, 8 including the systematic identification of genes with disputed or refuted relationships to disease.9 However, with over 3,000 genes reported in association with Mendelian disease,10 and many of these linked to multiple phenotypes, there is a significant unmet need for evidence-based curated gene resources that can be applied in diagnostic practice today.
Adding further complexity, a common strategy used to increase diagnostic efficiency is the collation of gene-disease validity assessments into “virtual” gene panels for specific testing indications such as intellectual disability. This approach targets genomic analysis to variants that are most likely to be clinically relevant while reducing secondary findings.11 However, it necessitates appropriate panel design, maintenance, and application to mitigate the risk of missing diagnoses, particularly for patients with complex, poorly defined, still evolving, or blended phenotypes. The wide variability in panel content between diagnostic providers is well recognized and raises significant concerns about impact on diagnostic and patient outcomes.12, 13, 14, 15 Although guidance on panel design and maintenance has been recently provided by the American College of Medical Genetics and Genomics (ACMG),16 there are few, if any, efforts to systematically harmonize these processes across healthcare systems,17 which results in variability in practice and duplication of effort.
Here, we describe our experience of using PanelApp, an open-access gene and panel curation platform, across two healthcare systems and multiple diagnostic providers with the aim of harmonizing gene panel content through systematic discordance resolution.
PanelApp background
PanelApp is an open-access platform developed to support virtual gene panel curation and incorporate crowd-sourced reviews from multiple experts worldwide.18 The application programming interface (API) can be integrated into sequencing analysis pipelines or variant curation platforms to assist in variant prioritization, and all panel content is downloadable. Gene-disease validity assessments are captured through structured reviews, including disease association, mode of inheritance, and supporting publications. The platform also enables curation of recurrent copy number variant sites (CNVs) and known short tandem repeat sites (STRs). PanelApp curators evaluate the reviews and current evidence to designate an overall rating to the gene-disease assessment. Three thresholds based on a traffic light system determine which genes have sufficient evidence to be analyzed for a clinical indication. The “green” rating denotes genes used in diagnostic reporting and requires case-level evidence from three unrelated families or two unrelated families with convincing functional data. The green PanelApp rating was developed from G2P and ClinGen guidelines specifically to be relevant for a wide range of rare diseases. “Amber” and “red” ratings reflect gene-disease associations with moderate or low levels of evidence, respectively. The PanelApp infrastructure is an open-source Python application under the Apache 2 license and is freely available with deployment support for commercial cloud vendors and a local environment for development and testing. The cloud version utilizes managed services wherever possible, reducing maintenance requirements and simplifying integration with existing computational infrastructure.
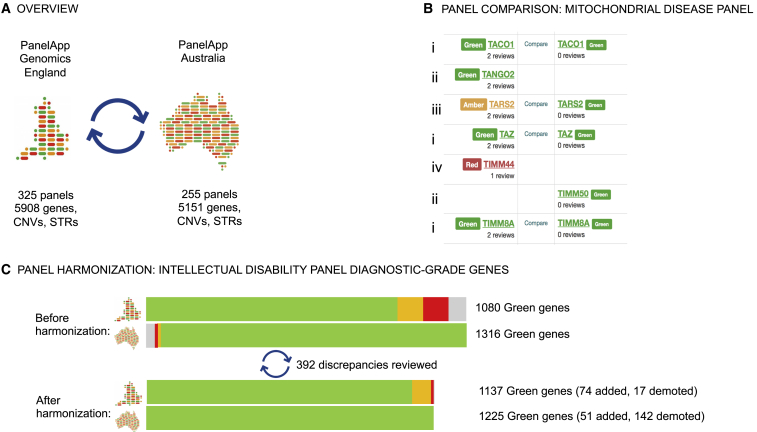
The platform was designed by Genomics England for the 174 rare disease and cancer susceptibility panels used in the 100,000 Genomes Project.19 Building upon this, the National Health Service (England) (NHSE) Genomic Medicine Service (GMS) is poised to deliver genomic testing through a network of genomic laboratory hubs (GLHs) for clinical indications defined in the National Genomic Test Directory. The Genomics England PanelApp instance has transitioned to support the GMS by harnessing clinical and laboratory knowledge from GLHs to enable expert curation of 173 panels for diagnostic use. Clinical validity assessments are currently available for 5,859 genes, 63 CNVs, and 21 STRs (Figure 1A).
Figure 1.
Virtual gene panel comparison and harmonization via PanelApp
(A) The PanelApp instances deployed in the UK and Australia use a common data model and curation guidelines but maintain separate governance and panel repertoires.
(B) The “compare panels” feature in PanelApp can be used to visualize discrepancies between gene panels, as displayed here for a section of the mitochondrial disease panels. The comparison tool highlighted gene rating agreement (i) as well as discrepancies requiring review, including diagnostic-grade genes included in one panel but not the other (ii) and genes with different ratings (iii). Red genes on one panel but missing on the other (iv) were not reviewed because of low level of evidence for inclusion.
(C) Improved alignment between the two intellectual disability panels following exchange of reviews. The green bars represent number of genes rated “green” on each panel. The gray, amber, and red bars represent genes that were rated “green” on one panel but were either absent (gray) or discordantly rated on the other.
PanelApp Australia was deployed in 2019 by the Australian Genomics Health Alliance, a national collaborative research partnership piloting approaches and developing evidence to inform national integration of genomics into mainstream care.20 It currently hosts 255 virtual gene panels developed by Australian genomics rare disease and cancer cohort studies, as well as panels created by diagnostic laboratories and clinical services. Clinical validity assessments are available for 5,105 genes, 62 CNVs, and 30 STRs. The Australian instance has been used for consolidation of virtual panels between Australian diagnostic services with the aim of national harmonization.
Maintaining separate PanelApp instances allows separate governance in relation to panel repertoire, which is influenced by local factors such as genomic test funding for specific clinical indications. It also supports autonomous decision-making and facilitates local accreditation and integration into diagnostic pipelines. For example, gene panel versions used for diagnosis in the NHSE are overseen by the NHSE Genomics Clinical Reference Group and reviewed by test evaluation working groups, while individual Australian diagnostic laboratories retain oversight of the panels they use. On the other hand, the use of the same underlying software enables technical co-development and pooling of curated data and expertise to drive quality improvements.
Panel comparison and discordance resolution
Comparing genetic variant pathogenicity classifications by different laboratories to identify discordance is a powerful tool for improving the quality of variant curation. It has been facilitated at scale by platforms such as ClinVar, where variant assessments are deposited by multiple providers via a structured format.21,22 The publication of widely adopted standardized criteria for assessing variant pathogenicity,23 and quality improvement schemes measuring concordance, such as those initiated by the Clinical Sequencing Evidence-Generating Research (CSER) Consortium,24,25 the European Molecular Genetics Quality Network (EMQN), and Genomics Quality Assessment (GenQA), which is part of the UK National External Quality Assessment Service (NEQAS) Consortium, have improved consistency between diagnostic providers. Similar efforts in gene curation are hampered by the lack of agreed terminology, agreed curation standards, and platforms that can be used to identify and resolve discrepancies at scale. The use of a common platform including a shared data model and gene curation scheme enabled us to undertake rapid systematic comparison between gene panels used in the UK and Australia with the aim of harmonizing content and identifying areas for process improvement.
Comparing approaches to panel design
First, we performed mapping to identify comparable gene panels (panel pairs) between the two PanelApp instances, which presented several immediate challenges. Gene panel names do not utilize disease ontologies but are named on the basis of common clinical indications for testing. Although the names are usually a good guide to content, panel comparability is also determined by the rules applied in panel design and by clinical context. For example, the two “photosensitivity disorders” panels had non-overlapping content: one contained genes primarily related to porphyrias and the other contained genes primarily related to DNA repair disorders. The “hearing loss” and “deafness” panels only partially overlapped in content because of the inclusion of syndromic disorders in one panel but not the other, and a similar issue was identified between the two “genetic epilepsy” panels: metabolic disorders were included in one panel but not the other. We also encountered significant differences in how panels are subdivided within disease categories. For example, PanelApp Australia contains 17 panels covering primary immunological disorders, whereas Genomics England PanelApp has a single panel. To enable comparison, we used the PanelApp “superpanels” functionality, which groups panels together, such as within a disease category (i.e., immunological disorders).
The mapping exercise highlighted common tensions in panel design in relation to limiting or broadening panel scope14 on the basis of factors such as age of presentation (e.g., adult-onset ataxia) or absence of syndromic features (e.g., non-syndromic hearing loss). Smaller panels can widen access to genomic testing by enabling sub-specialists to order genomic tests within their area of expertise. From a laboratory perspective, smaller panels increase clinical specificity while limiting the number of variants of uncertain significance. However, this approach carries diagnostic risks if limited panels are applied to investigate patients with complex, evolving, and blended phenotypes. Further, smaller panels can add complexity to test requisition and can act as a barrier for mainstream clinicians. The two PanelApp instances increasingly support overarching panels or superpanels within and across disease categories (“kidneyome,” “pediatric disorders,” “Mendeliome”) to facilitate comprehensive analysis for complex patients and aid the transition of genomic testing toward a first-tier mainstream test used early in the diagnostic trajectory. These shifts need to be coupled with education, training, and decision support tools to promote best practice as part of test requisition, analysis, and reporting. Mapping of panel clinical indications to disease ontologies such as the Monarch Disease Ontology (MONDO)26 will assist in both harmonization and automation efforts. Most importantly, the rules governing panel design and use need to be articulated clearly to guide clinicians and diagnostic laboratories, particularly because open resources like PanelApp have a wide range of international users (Box 1, key recommendations).
Box 1. Key recommendations for the development, use, and maintenance of virtual gene panels.
-
•
Use a standardized gene curation framework during panel development and set clear evidence thresholds for diagnostic use
-
•
Use standard ontologies and record case-level and functional evidence for each gene-disease association and proposed mode of inheritance
-
•
Provide clear panel descriptions to guide clinicians and laboratories in panel application, explaining the clinical indication and listing any significant clinical or technical exclusions
-
•
Encourage application of multiple panels (co-application) and the use of larger panels within and across disease categories to mitigate the risks of restricted analysis
-
•
Make your gene-disease and virtual gene panel data openly available and promote collaboration and harmonization nationally and internationally to reduce duplication of effort and improve diagnostic outcomes
-
•
Focus expert gene curation effort on resolving discrepancies and on continuous surveillance and assessment of new evidence for gene-disease associations to enable rapid gains in evidence-based practice
-
•
Use version-controlled panels and provide easy access to previous versions to ensure a clear understanding of the analysis performed and guide future re-analysis
-
•
Develop specific training and competencies in gene and virtual panel curation across the genomic workforce and enable wide participation in the curation effort
Comparing and harmonizing panel content
We identified 80 comparable panel pairs and, allowing for differences arising from panel scope definition, reviewed their content to identify discrepancies arising from differences in gene-disease validity assessment and relevance to panel indication. We uploaded panels from the Genomics England instance to the Australian instance in order for the panels to be compared side-by-side with the “compare panels” functionality in PanelApp; visualizing panels side-by-side readily identifies discrepant gene pairs. This includes genes that are present on one panel but absent from the other and genes with discrepant ratings, for example, “green-red” (Figure 1B). Genes rated red in one instance but absent from the other were not assessed. All discrepant gene pairs were reviewed initially by the Australian curation team, assessing the evidence provided by the Genomics England curation team. Where the Australian team disagreed, a review with the additional evidence was provided on the Genomics England PanelApp instance. These reviews were assessed independently by the Genomics England PanelApp curation team, resulting in further discrepancy resolution.
Of the 80 panel pairs compared, only three relatively small panels covering highly specific clinical indications were completely concordant (hematuria, osteopetrosis, and chondrodysplasia punctata). A total of 2,144 discrepant gene ratings and inclusions were identified in the remaining 77 panel pairs. The evidence for gene-disease association and relevance to panel clinical indication was assessed for each discrepancy, and structured reviews were exchanged between the two curation teams via the PanelApp platforms for the promotion of discordance resolution. Of the reviews exchanged, 1,008 reviews (47%) indicated that there was sufficient evidence for diagnostic use (green rating) and 1,139 reviews (53%) indicated moderate or limited evidence for gene-disease association or panel inclusion (425 amber and 714 red). Common sources of discordance are presented in Table S1, and all reviews exchanged and curator assessments are publicly available on the PanelApp instances.
As an example, the largest panel, intellectual disability (ID), had the largest number of discrepancies, 595, including 392 affecting diagnostic-grade (green) genes. Of these, 39 were due to a difference in panel design and were not reviewed further: the Genomics England PanelApp panel contains genes associated with moderate to profound ID, and complex disorders are included if ID is a presenting feature. Individuals presenting predominantly with other clinical features can access testing through alternative pathways and their associated virtual panels (e.g., inborn errors of metabolism) because genomic testing is funded through the healthcare system for a range of indications. By contrast, funding of genomic testing in Australia is more variable; federal funding is currently only available for syndromic and non-syndromic ID, necessitating more inclusive panel design. Following re-evaluation of the evidence for each of the remaining 353 discordances involving diagnostic-grade (green) genes, 298 (84%) were resolved (Figure 1C). Fifty-five (16%) remained discordant; all were rated green in PanelApp Australia and amber/red in Genomics England PanelApp. The most common reasons for remaining discrepant ratings were variability in the level of ID reported (22), insufficient cases to determine whether ID is a key presenting feature (17), and differential weighing of functional evidence where only two unrelated families are reported in the literature (9).
Building the evidence base for gene-disease associations
The pace of rare disease gene discovery continues unabated,27 presenting a constant challenge for curated databases of gene-disease associations to maintain currency with flow-on effects on diagnostic outcomes both for analysis of new data and reanalysis of existing data.28,29 The ACMG recommends regular monitoring and revision of diagnostic panels and at least a 6-monthly review cycle incorporating consultation with disease experts, literature surveys, and monitoring of curated databases.16 We combined the curator resources across the two PanelApp instances to conduct a monthly literature review of 20 genetics and other subspecialty journals (Table S2) to augment crowdsourced expert reviews and monitoring of other curated databases. Over the first 6 months, this process resulted in 219 new gene curations. This included 157 novel gene-disease associations and 62 changes to existing gene curations incorporating reports of novel allelic disorders, phenotype expansions, new modes of inheritance, or additional case reports or functional data altering evidence ratings.
A total of 107 new gene-disease associations with enough evidence for diagnostic use were identified between June and November 2020. Of these, 41 (38%) are yet to be included in OMIM, highlighting the importance and value of a proactive approach to the identification and curation of new evidence for diagnostic benefit. We found frequent, regular searches for new evidence easier to integrate with existing workflows, while the pooling of curator resources on an open platform reduced individual curation burden. However, the optimal frequency of panel updates needs to take governance and other laboratory requirements into consideration; some diagnostic providers may elect to integrate new information into diagnostic pipelines at less frequent intervals, for example quarterly or 6-monthly, by utilizing panel version control features.
Gene curation and panel harmonization: Future directions
The harmonization effort described here has several, chiefly technical, limitations highlighting the need for ongoing platform development.30 Our approach relied on manual transfer of reviews, which is laborious; automating key notifications between PanelApp instances, for example when genes are promoted to or demoted from a diagnostic rating (green), would increase efficiency and benefit the wider community. The use of literature mining tools to identify reports of gene-disease associations would make the new gene identification effort more comprehensive while allowing gene curators to focus on critical appraisal. Most of the work undertaken to date relates to genes implicated in rare Mendelian disorders, and further co-development should address the broader applications of diagnostic and research genomics, including somatic cancer testing and complex polygenic disorders.
In conclusion, maintaining curated gene-disease association databases for diagnostic use is a complex task and benefits from pooling expertise across multiple professional groups, including diagnostic scientists, bioinformaticians, clinicians, and researchers. The necessity for broad input has to be coupled with expertise in gene curation, and we need to develop resources, provide training, and establish competencies across the genomic workforce. Sharing interpretation knowledge and diagnostic experience across multiple providers, including internationally, is key to accelerating the implementation of genomics in healthcare.31
We have demonstrated the utility of a collaborative approach to identifying and resolving discrepancies in gene and virtual gene panel curation facilitated by the use of a common data platform. The PanelApp software is open source and available for other healthcare systems to implement and participate in data sharing. Harmonization of gene-disease validity assessments between gene curation initiatives will be facilitated by the development of agreed terminology, federated platforms for sharing knowledge via standardized formats, and a focused curation effort to resolve discrepancies. We anticipate that the establishment of the Gene Curation Coalition, of which both Genomics England PanelApp and PanelApp Australia are members, will serve as a focal point for international harmonization efforts. The large number of discrepancies resolved here highlights the scale of the challenge but also provides a blueprint for extending this effort in the future.
Acknowledgments
We thank all PanelApp reviewers and those who have contributed feedback or gene lists to help in the development of PanelApp; individual panels show the names and affiliations of contributors. The Australian Genomics Health Alliance is supported by the Australian National Health and Medical Research Council (GNT1113531). M.J.C. was funded by the National Institute for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Center at Barts and The London School of Medicine and Dentistry. He is supported as an NIHR senior investigator, and this work was funded by the MRC eMedLab award. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health). The 100,000 Genomes Project is funded by the NIHR and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the NHS England as part of their care and support. We thank all participants in the 100,000 Genomes Project.
Declaration of interests
Kathryn North is a member of the Editorial Board of The American Journal of Human Genetics. The other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.020.
Web resources
ClinGen, https://clinicalgenome.org
European Molecular Genetics Quality Network (EMQN), https://www.emqn.org
Gene2Phenotype, https://www.ebi.ac.uk/gene2phenotype
Gene Curation Coalition, https://thegencc.org/
Genomics England PanelApp, https://panelapp.genomicsengland.co.uk/
Genomics Quality Assessment (GenQA), https://www.genqa.org/
NHS England National Genomic Test Directory, https://www.england.nhs.uk/publication/national-genomic-test-directories/
OMIM, https://omim.org/
PanelApp Australia, https://panelapp.agha.umccr.org/
PanelApp GitLab, https://gitlab.com/genomicsengland/panelapp/panelapp
United Kingdom National External Quality Assessment Service (UK NEQAS), https://ukneqas.org.uk
Supplemental information
References
- 1.Thormann A., Halachev M., McLaren W., Moore D.J., Svinti V., Campbell A., Kerr S.M., Tischkowitz M., Hunt S.E., Dunlop M.G. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat. Commun. 2019;10:2373. doi: 10.1038/s41467-019-10016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strande N.T., Riggs E.R., Buchanan A.H., Ceyhan-Birsoy O., DiStefano M., Dwight S.S., Goldstein J., Ghosh R., Seifert B.A., Sneddon T.P. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017;100:895–906. doi: 10.1016/j.ajhg.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler A., Novelli V., Amin A.S., Abiusi E., Care M., Nannenberg E.A., Feilotter H., Amenta S., Mazza D., Bikker H. An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation. 2020;141:418–428. doi: 10.1161/CIRCULATIONAHA.119.043132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGlaughon J.L., Pasquali M., Wallace K., Ross J., Senol-Cosar O., Shen W., Weaver M.A., Feigenbaum A., Lyon E., Enns G.M. Assessing the strength of evidence for genes implicated in fatty acid oxidation disorders using the ClinGen clinical validity framework. Mol. Genet. Metab. 2019;128:122–128. doi: 10.1016/j.ymgme.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 5.DiStefano M.T., Hemphill S.E., Oza A.M., Siegert R.K., Grant A.R., Hughes M.Y., Cushman B.J., Azaiez H., Booth K.T., Chapin A. ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet. Med. 2019;21:2239–2247. doi: 10.1038/s41436-019-0487-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingles J., Goldstein J., Thaxton C., Caleshu C., Corty E.W., Crowley S.B., Dougherty K., Harrison S.M., McGlaughon J., Milko L.V. Evaluating the Clinical Validity of Hypertrophic Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2019;12:e002460. doi: 10.1161/CIRCGEN.119.002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant A.R., Cushman B.J., Cavé H., Dillon M.W., Gelb B.D., Gripp K.W., Lee J.A., Mason-Suares H., Rauen K.A., Tartaglia M. Assessing the gene-disease association of 19 genes with the RASopathies using the ClinGen gene curation framework. Hum. Mutat. 2018;39:1485–1493. doi: 10.1002/humu.23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renard M., Francis C., Ghosh R., Scott A.F., Witmer P.D., Adès L.C., Andelfinger G.U., Arnaud P., Boileau C., Callewaert B.L. Clinical Validity of Genes for Heritable Thoracic Aortic Aneurysm and Dissection. J. Am. Coll. Cardiol. 2018;72:605–615. doi: 10.1016/j.jacc.2018.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosseini S.M., Kim R., Udupa S., Costain G., Jobling R., Liston E., Jamal S.M., Szybowska M., Morel C.F., Bowdin S. Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation. 2018;138:1195–1205. doi: 10.1161/CIRCULATIONAHA.118.035070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthijs G., Souche E., Alders M., Corveleyn A., Eck S., Feenstra I., Race V., Sistermans E., Sturm M., Weiss M. Guidelines for diagnostic next-generation sequencing. Eur. J. Hum. Genet. 2016;24:1515. doi: 10.1038/ejhg.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers C., Jansen L.A., Dhamija R. Review of Commercially Available Epilepsy Genetic Panels. J. Genet. Couns. 2016;25:213–217. doi: 10.1007/s10897-015-9906-9. [DOI] [PubMed] [Google Scholar]
- 13.Pekeles H., Accogli A., Boudrahem-Addour N., Russell L., Parente F., Srour M. Diagnostic Yield of Intellectual Disability Gene Panels. Pediatr. Neurol. 2019;92:32–36. doi: 10.1016/j.pediatrneurol.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Pellacani S., Dosi C., Valvo G., Moro F., Mero S., Sicca F., Santorelli F.M. Customized multigene panels in epilepsy: the best things come in small packages. Neurogenetics. 2020;21:1–18. doi: 10.1007/s10048-019-00598-x. [DOI] [PubMed] [Google Scholar]
- 15.Erger F., Schaaf C.P., Netzer C. Which genes to assess in the NGS diagnostics of intellectual disability? The case for a consensus database-driven and expert-curated approach. Mol. Cell. Probes. 2019;45:84–88. doi: 10.1016/j.mcp.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Bean L.J.H., Funke B., Carlston C.M., Gannon J.L., Kantarci S., Krock B.L., Zhang S., Bayrak-Toydemir P., ACMG Laboratory Quality Assurance Committee Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2020;22:453–461. doi: 10.1038/s41436-019-0666-z. [DOI] [PubMed] [Google Scholar]
- 17.Krahn M., Biancalana V., Cerino M., Perrin A., Michel-Calemard L., Nectoux J., Leturcq F., Bouchet-Séraphin C., Acquaviva-Bourdain C., Campana-Salort E. A National French consensus on gene lists for the diagnosis of myopathies using next-generation sequencing. Eur. J. Hum. Genet. 2019;27:349–352. doi: 10.1038/s41431-018-0305-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin A.R., Williams E., Foulger R.E., Leigh S., Daugherty L.C., Niblock O., Leong I.U.S., Smith K.R., Gerasimenko O., Haraldsdottir E. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat. Genet. 2019;51:1560–1565. doi: 10.1038/s41588-019-0528-2. [DOI] [PubMed] [Google Scholar]
- 19.Turnbull C., Scott R.H., Thomas E., Jones L., Murugaesu N., Pretty F.B., Halai D., Baple E., Craig C., Hamblin A. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 20.Stark Z., Boughtwood T., Phillips P., Christodoulou J., Hansen D.P., Braithwaite J., Newson A.J., Gaff C.L., Sinclair A.H., North K.N. Australian Genomics: A Federated Model for Integrating Genomics into Healthcare. Am. J. Hum. Genet. 2019;105:7–14. doi: 10.1016/j.ajhg.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison S.M., Dolinksy J.S., Chen W., Collins C.D., Das S., Deignan J.L., Garber K.B., Garcia J., Jarinova O., Knight Johnson A.E. Scaling resolution of variant classification differences in ClinVar between 41 clinical laboratories through an outlier approach. Hum. Mutat. 2018;39:1641–1649. doi: 10.1002/humu.23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison S.M., Dolinsky J.S., Knight Johnson A.E., Pesaran T., Azzariti D.R., Bale S., Chao E.C., Das S., Vincent L., Rehm H.L. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet. Med. 2017;19:1096–1104. doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amendola L.M., Muenzen K., Biesecker L.G., Bowling K.M., Cooper G.M., Dorschner M.O., Driscoll C., Foreman A.K.M., Golden-Grant K., Greally J.M. Variant Classification Concordance using the ACMG-AMP Variant Interpretation Guidelines across Nine Genomic Implementation Research Studies. Am. J. Hum. Genet. 2020;107:932–941. doi: 10.1016/j.ajhg.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;99:247. doi: 10.1016/j.ajhg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mungall C.J., McMurry J.A., Köhler S., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2017;45(D1):D712–D722. doi: 10.1093/nar/gkw1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamshad M.J., Nickerson D.A., Chong J.X. Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am. J. Hum. Genet. 2019;105:448–455. doi: 10.1016/j.ajhg.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright C.F., McRae J.F., Clayton S., Gallone G., Aitken S., FitzGerald T.W., Jones P., Prigmore E., Rajan D., Lord J. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med. 2018;20:1216–1223. doi: 10.1038/gim.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark Z., Schofield D., Martyn M., Rynehart L., Shrestha R., Alam K., Lunke S., Tan T.Y., Gaff C.L., White S.M. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet. Med. 2019;21:173–180. doi: 10.1038/s41436-018-0006-8. [DOI] [PubMed] [Google Scholar]
- 30.Knowles R.M., Mateen B.A., Yehudi Y. We need to talk about the lack of investment in digital research infrastructure. Nat. Comput. Sci. 2021 doi: 10.1038/s43588-021-00048-5. Published online March 25, 2021. [DOI] [PubMed] [Google Scholar]
- 31.Stark Z., Dolman L., Manolio T.A., Ozenberger B., Hill S.L., Caulfied M.J., Levy Y., Glazer D., Wilson J., Lawler M. Integrating Genomics into Healthcare: A Global Responsibility. Am. J. Hum. Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.