Figure 1.
mRNA splicing and sequence logos/walkers
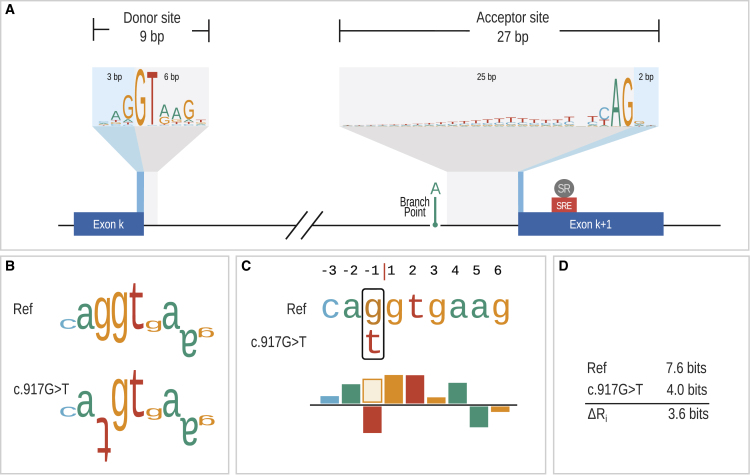
(A) The figure shows an intron and the corresponding canonical splice donor and acceptor sites, which are represented as logos, where the letters representing the sequence are stacked on top of each other for each position in the splice site. The height of the character stack at each position represents the sequence information gained, Rsequence(l), by aligning wild-type sequences of exon/intron junctions of GENCODE transcripts (material and methods). The heights of the characters within a stack represent contributions of the individual bases to the position.
(B) Individual sequence information (Ri) for a wild-type splice donor sequence of CHRNE (MIM: 100725) and for the corresponding sequence with the variant GenBank: NM_000080.3; c.917G>T (p.Arg306Met). c.917G>T is located at the last (3′ most) position of an exon and although it is predicted to lead to a missense change, it reduces the strength of the donor sequence and leads to skipping of the affected exon.46 The sequence walker representations as introduced by Schneider38 are shown for the wild-type and variant sequences. Sequence walkers display nucleotides that represent favorable contacts to the spliceosome and a test sequence by letters that extend upward and positions that are predicted to make unfavorable contacts are shown by inverted letters.
(C) SQUIRLS introduces a new graphical representation in which a bar chart is used to show the degree to which a sequence “matches” the donor or acceptor model. The height of the bars is calculated in the same way as for the height of the letters in the sequence walker. Positions that are changed by a variant are displayed such that the original nucleotide is shown as an outline (the “g” in this example) and the variant (alternate) base is shown filled.
(D) The variant reduces the Ri from 7.6 to 4.0 bits. Changes in Ri are referred to as . SQUIRLS calculates in several contexts (Figures 2 and 3).