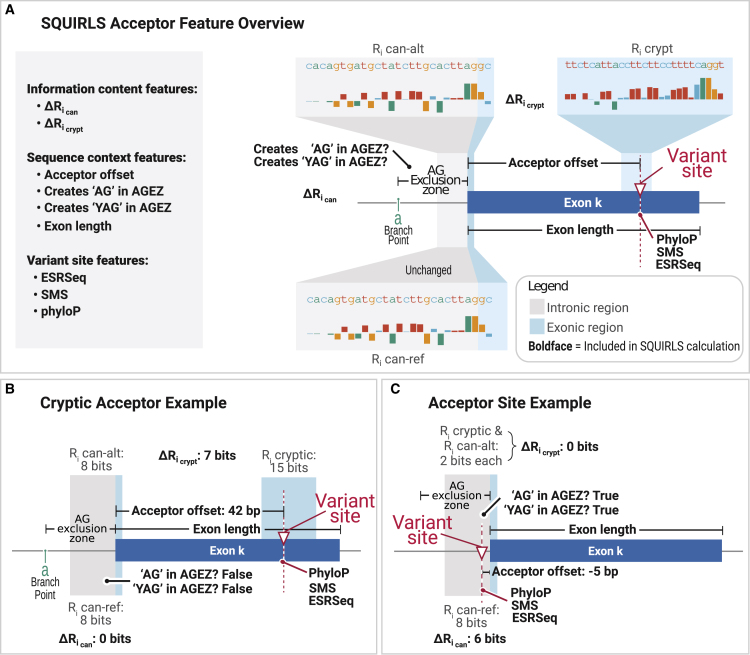
Figure 3.
Calculation of features for the splice acceptor site
(A) SQUIRLS calculates as the difference in the Ri between the reference and alternate sequence of the canonical acceptor sequence (Figure 1A). If the variant is located outside of this sequence, = 0. SQUIRLS evaluates the potential of variants to create cryptic splice sites using a sliding window approach (material and methods). is calculated by subtracting Ri of the alternate sequence of the canonical acceptor site from the Ri of the best candidate cryptic splice site. The random forest for acceptor variants does not use as our initial analysis showed that it did not boost classification performance. See Table 1 for information about other features.
(B) In this example, a coding variant activates a cryptic acceptor site with 15 bits which is greater than the canonical acceptor site by 7 bits ().
(C) A situation where a variant located in a splice acceptor site introduces novel AG dinucleotide into the AGEZ leading to cryptic splicing or exon skipping.