Figure 5.
Performance of SQUIRLS, SpliceAI, S-CAP, CADD, and MaxEnt on non-canonical SAVs
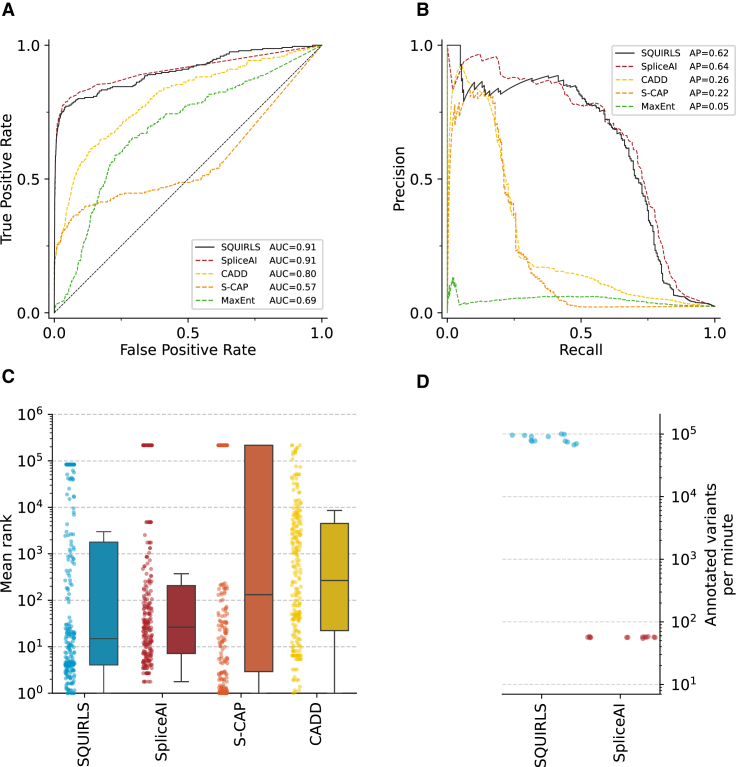
(A) Receiver operating characteristic curves indicate that SQUIRLS and SpliceAI achieve comparable performance.
(B) Precision-recall curves show that SQUIRLS and SpliceAI are able to find the most of the true splice variants, while maintaining high precision.
(C) Mean ranks of splice variants among background variants in simulated exome sequencing runs. Each dot represents the mean rank of one of the splice variants in 13 WES simulations. The boxes represent distributions of the mean ranks. The horizontal line of each box indicates the median mean rank, box borders indicate positions of the 1st and the 3rd quartile, and the whiskers indicate 1.5× the interquartile range.
(D) Comparison of algorithm runtimes for SQUIRLS and SpliceAI. We recorded the time required for analysis of 13 VCF files containing 87,000–107,000 variants. The figure shows the annotation speed that was achieved on a consumer laptop. We could not compare the performance of S-CAP and CADD, since they provide precomputed predictions as tabular files. Therefore, the annotation speed is only dependent on a package used to query the tabular file (e.g., tabix).