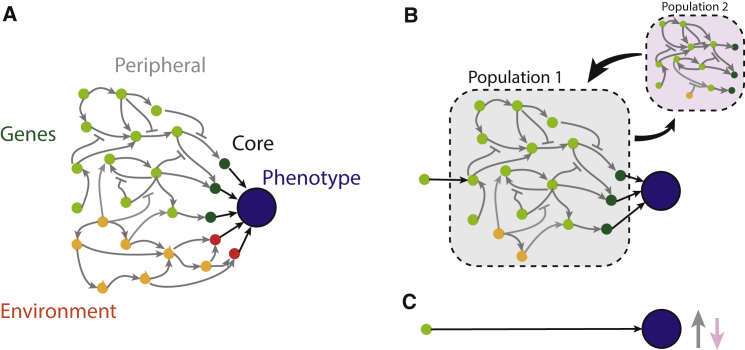
Figure 1.
The omnigenic model
(A) Schematic of the omnigenic model (after Liu et al.4). Genetic factors in green and environmental factors in orange, with arrows showing interactions. Peripheral factors are lightly shaded whereas core factors with direct effects on phenotype are darkly shaded.
(B) A gene’s-eye perspective. A single peripheral gene’s effect on the phenotype is filtered through part of the network (gray box).
(C) The GWAS perspective. The effect of a causal variant as measured by an association study in a single population (gray arrow in C) is the expected value of its effect, with respect to the distribution of genetic (i.e., allele frequencies) and environmental factors in the population. In a different population (pink box in B), the weights and possibly structure of the network change, leading to a different expected effect size (pink arrow in C).