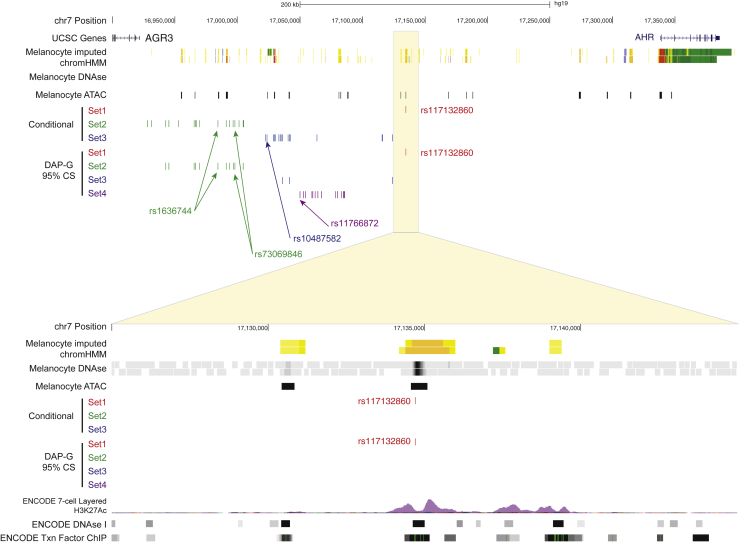
Figure 1.
Fine-mapping of multiple melanoma GWAS signals on chromosome band 7p21.1
(Top) A view of the 7p21.1 locus including UCSC genes, imputed ChromHMM and DNaseI hypersensitivity (DHS) data from two melanocyte cultures generated by the RoadMap Project, ATAC-seq data generated from five human primary melanocyte cultures, and candidate causal variant sets nominated by either conditional analysis or Bayesian fine-mapping (95% credible sets for each of four clusters) with DAP-G. Candidate causal variants mapping to signal/cluster 1 are in red, those mapping to signal/cluster 2 are in green, those mapping to signal/cluster 3 are in blue, and those mapping to signal/cluster 4 are in purple. (Bottom) A zoomed-in view of the region immediately surrounding rs117132860, showing rs117132860 mapping to open chromatin and annotated melanocyte enhancer. Genomic positions are based on hg19. For the zoomed-in region containing rs117132860 (bottom), layered 7-cell H3K27Ac, DHS clusters, and transcription factor CHIP tracks were generated by ENCODE and provided through the UCSC Genome Browser.