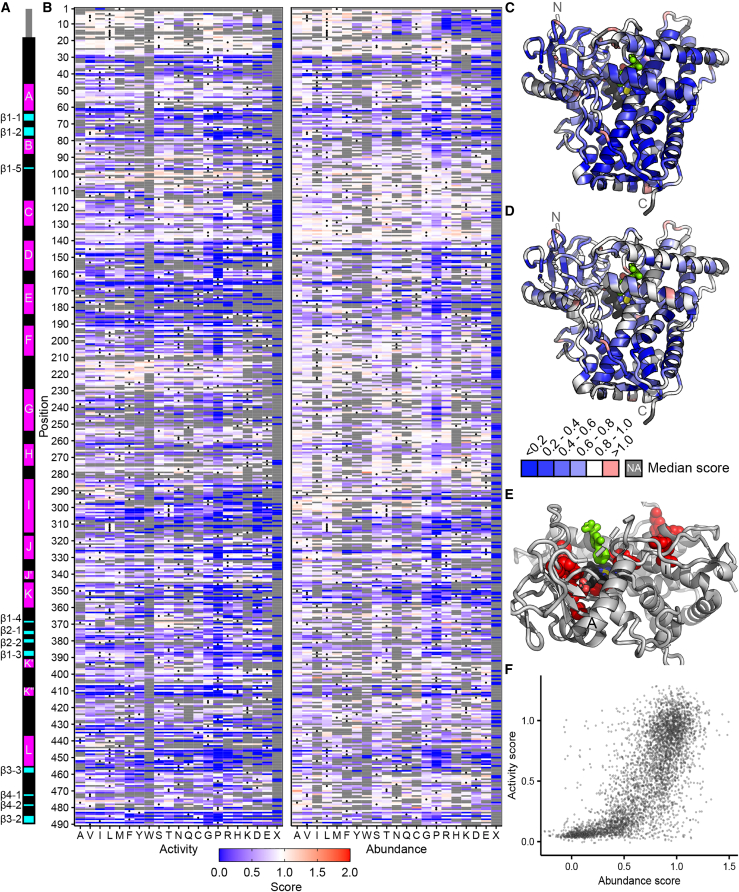
Figure 4.
Click-seq activity scores and VAMP-seq abundance scores for CYP2C9
(A) Secondary structure of CYP2C9; alpha helices are shown in magenta, and beta sheets are shown in cyan. Helix and beta sheet names are labeled.
(B) Heatmaps of CYP2C9 activity (left) and abundance (right) scores. WT amino acids are denoted with a dot, and missing data are shown in gray. Scores range from nonfunctional (blue) to WT-like (white) to increased (red).
(C and D) CYP2C9 structure (PDB: 1R9O) colored by median activity (C) and abundance (D) at each position. Median scores are binned as depicted in the legend, and missing positions are shown in gray. Heme is colored by element (carbon:black, nitrogen:blue, oxygen:red, iron:yellow), and substrate (flurbiprofen) is colored bright green. Median activity scores shown in (C), and median abundance scores shown in (D).
(E) Zoomed view of partial CYP2C9 structure. Positions with the lowest 2.5% specific activity scores are shown as red spheres. F and G helices are hidden, A and I helices are labeled, and heme and substrate are colored as in (C) and (D).
(F) Scatterplot of CYP2C9 activity and abundance scores from a total of 4,421 missense variants.