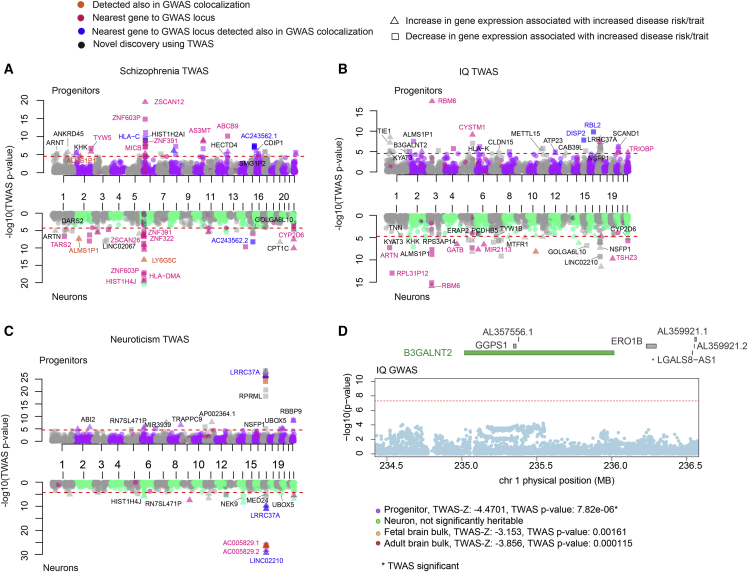
Figure 6.
Prediction of differential gene expression during human brain development via TWAS
(A) Manhattan plots for schizophrenia, IQ, and neuroticism TWAS for progenitors (purple-gray, top) and neurons (green-gray, bottom) where the LD matrix used was based on a European population. Each dot shows −log10 (TWAS p value) for each gene on the y axis, gene names were color coded based on discovery also in colocalization analysis (orange), defined as the nearest gene to GWAS locus (dark pink), being in both these two categories (blue), and discovered only in TWAS analysis (black). Only joint independent genes are labeled (positively and negatively correlated genes represented by triangle and square, respectively, and red line used for TWAS significant threshold).
(B) Manhattan plots for IQ TWAS, as described in (A).
(C) Manhattan plots for neuroticism TWAS, as described in (A).
(D) IQ TWAS results for B3GALNT2, regional association of variants to IQ trait shown at the top, and statistics from each TWAS study shown at the bottom (red line used for genome-wide significant threshold 5 × 10−8).