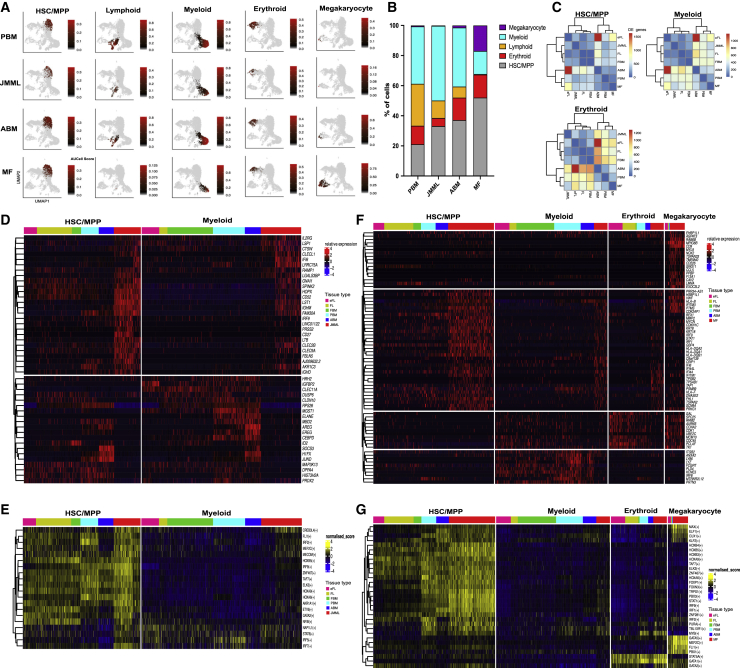
Figure 6.
Concordance in gene expression between juvenile myelomonocytic leukemia (JMML) HSPCs and normal fetal HSPCs, and increased inflammatory signaling seen in myelofibrosis (MF) and ABM in HSPCs
Data were derived from JMML (n = 2, total cells = 19,524) and MF (n = 15, total cells = 19,524) HSPCs.
(A) UMAP plots showing lineage progenitors as identified by AUCell score >0.15 for downsampled datasets to show 5,600 cells per tissue type.
(B) The proportion of HSPCs (from all of the cells within each tissue), classified as HSC/MPP or lineage progenitors in JMML, MF, and their normal counterparts.
(C) Hierarchical clustering of the number of differentially expressed genes (DEGs) from pairwise comparisons of tissues from normal ontogeny and JMML and MF for HSC/MPP, myeloid and erythroid compartments (adjusted p < 0.05, absolute log2FC > 1.5, and expressing cell frequency >0.3 per tissue).
(D and E) Top DEGs and regulons, respectively, between JMML HSPCs and those from normal human ontogeny tissues for HSC/MPP and myeloid progenitors.
(F and G) Comparison of top DEGs and regulons, respectively, between MF HSPCs and those from normal human tissues through ontogeny for HSC/MPP and myeloid, erythroid, and megakaryocyte lineage progenitors.