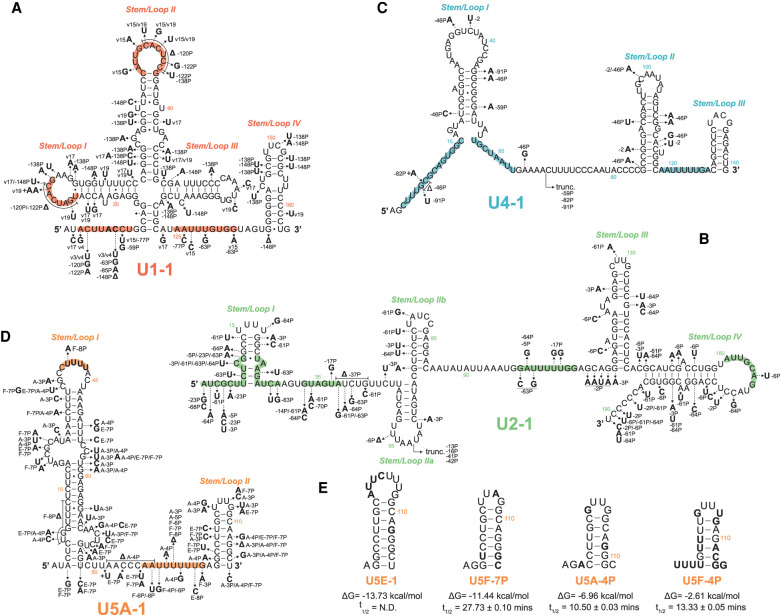
FIGURE 3.
Expressed snRNA variant genes exhibit sequence variation in RNA:RNA and RNA:protein interacting domains. Arrows show the positions of selected nucleotide variants relative to the canonical snRNA. Bold letters show the substituted base, followed by the variant snRNA(s) in which the substitution occurs. Colored regions within each structure are known functional sequences further described in Supplemental Figure 5. snRNA genes: (A) U1, (B) U2, (C) U4, (D) U5. (E) Predicted 3′ stem–loop structure of U5 snRNA variants with conserved or divergent base changes. Bold letters show the substituted base as compared to U5A. The free energy of formation (ΔG) and snRNA half-lives (t1/2) are shown below each snRNA variant structure. For reference, the free energy of formation for the 3′ stem of U5A-1 is −13.53 kcal/mol. Structures and free energy of folding were calculated using the Vienna RNAfold webserver (rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).