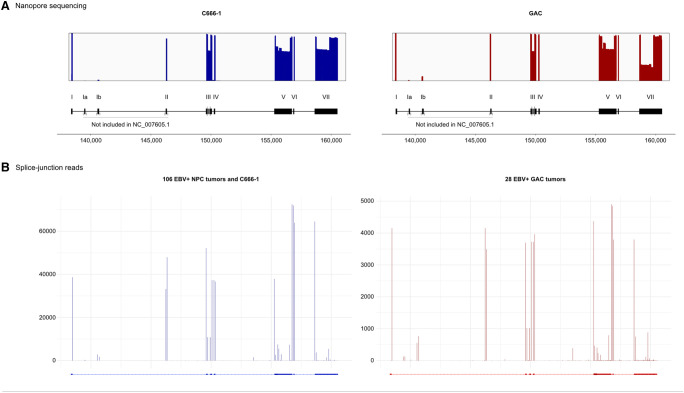
FIGURE 2.
Alignment of long nanopore sequencing reads and splice junctions in corresponding short-read sequencing data sets. (A) Coverage plots of nanopore sequencing reads across RPMS1 in C666-1 and GAC. Bars display alignment to constitutive (I–VII) and cassette exons (Ia and Ib), and numbers below refer to position in the EBV genome. Changes in coverage peaks correspond to splicing. The current RefSeq annotation (NC_007605.1) does not include exon Ia, Ib, and II, nor the multitude alternative splice sites within exon III, V, and VII. (B) Detection of splice junction in short-read sequencing data sets. The number in the y-axis corresponds to the frequency of splice-junction reads. The coverage of nanopore reads across RPMS1 correlates with the extracted splice-junction positions in complementary short-read sequencing data sets. (NPC) Nasopharyngeal carcinoma.