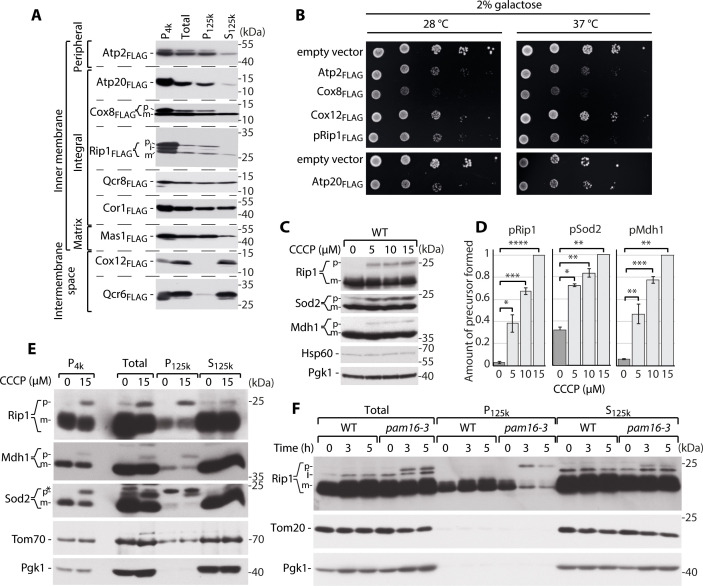
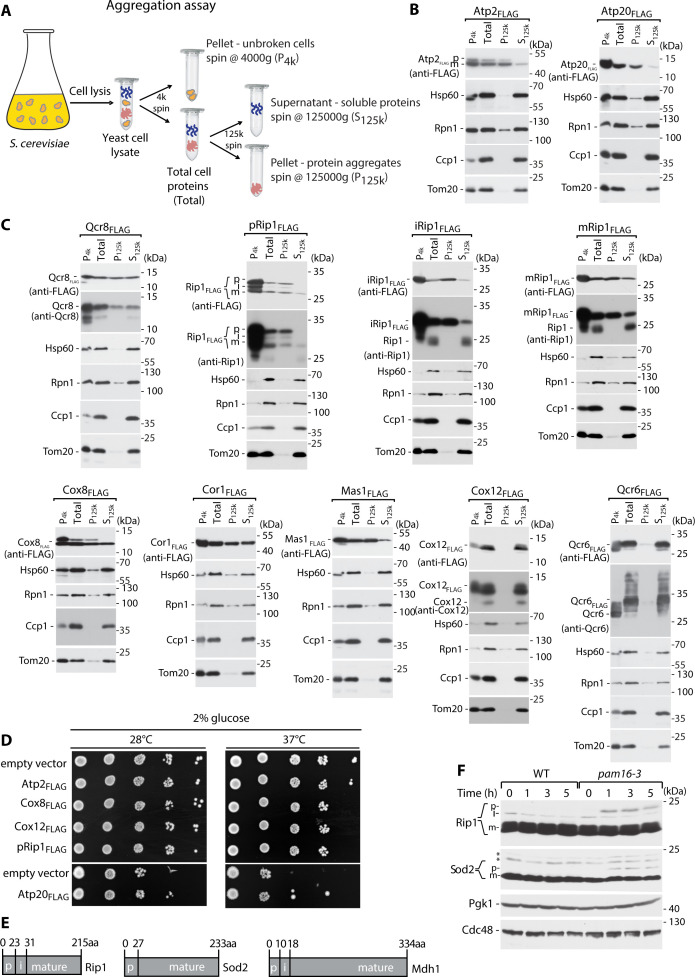
Figure 1. Supersaturated nuclear-encoded mitochondrial proteins aggregate in the cytosol.
(A) SDS-PAGE analysis of aggregation assay fractions of WT yeast cells that overexpressed Atp2FLAG, Atp20FLAG, Cox8FLAG, pRip1FLAG, Qcr8FLAG, Cor1FLAG, Mas1FLAG, Cox12FLAG, and Qcr6FLAG for 3 hr when 2% galactose with 0.1% glucose was used as the carbon source. Pellet fractions after 4000 and 125,000× g centrifugation are indicated as P4k and P125k, respectively. The soluble fraction at 125,000× g is indicated as S125k. n = 3. (B) Ten-fold dilutions of WT cells that expressed metastable proteins or controls that were spotted on selective medium agar plates with galactose as the main carbon source at 28 and 37°C. (C) Total protein cell extract from WT yeast grown at 24°C and treated with 0, 5, 10, or 15 µM carbonyl cyanide m-chlorophenyl hydrazine (CCCP) for 30 min. (D) Quantification of pRip1, pSod2, and pMdh1 from (C). Quantified data are shown as mean ± SEM. n = 3. (E) SDS-PAGE analysis of aggregation assay fractions of yeast cells that were treated with 15 µM CCCP for 30 min, with 2% sucrose as the carbon source. (F) SDS-PAGE analysis of aggregation assay fractions of WT (pam16WT) and pam16-3 mutant yeast strains grown at 19°C and shifted to 37°C for 0, 3, or 5 hr, with 2% sucrose as the carbon source. In (A), (C), (E), and (F), the samples were separated by SDS-PAGE and identified by western blot with specific antisera. n = 3 for each experiment. *Nonspecific; p: presequence protein; i: intermediate protein; m: mature protein. *p<0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.