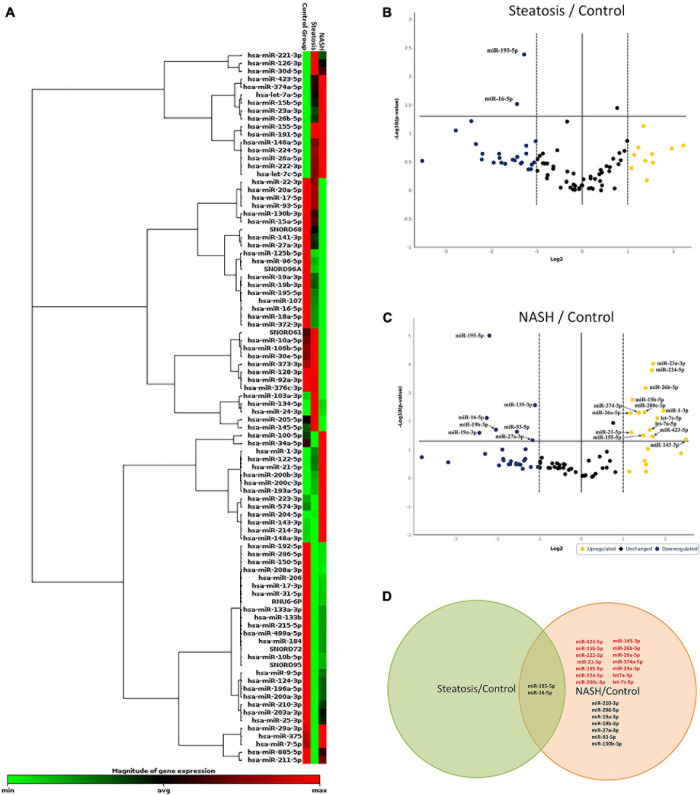
FIGURE 3.
Identification of differentially expressed miRNAs. (A) Hierarchical cluster analysis. Red—increased miRNA; green—decreased miRNA; black—no changes in expression. The magnitude of gene expression is determined by calculating the 2-ΔCT for each gene and normalizing to the average 2-ΔCT of all genes across all arrays. Volcano plot of differentially expressed miRNAs in serum; (B) group of patients with steatosis and (C) group of patients with NASH. Plots show the relationship between fold change and statistical significance. The solid vertical line shows no change in gene expression (log21 = 0). Data points to the right of the solid vertical line indicate activated genes, and data points to the left indicate repressed genes. The dashed lines represent the threshold—2. The solid vertical line represents the threshold of statistical significance, p < 0.05. Thus, the blue and yellow dots in the graph indicate >2.0-fold activation and suppression of expression with statistical significance. (D) Venn plot of miRNA profile overlap showing common and unique miRNAs in serum from groups of patients with steatosis and NASH. Expression activation is highlighted in red, while statistically significant expression suppression is highlighted in black (D).