Abstract
Purpose
Investigate the contribution of the Wnt pathway to vascular endothelial growth factor (VEGF)/anti-VEGF-mediated control of endothelial cell permeability.
Methods
High glucose-treated primary human retinal endothelial cells (HRECs) were exposed to either VEGF, or VEGF and then anti-VEGF. Changes in gene expression were assayed by RNAseq and qRT-PCR. Permeability was monitored by electrical cell-substrate impedance sensing (ECIS). Approaches to activate the Wnt pathway included treatment with LiCl and overexpression of constitutively activated β-catenin. β-catenin-dependent transcriptional activity was monitored in HRECs stably expressing a TCF/LEF-driven reporter.
Results
VEGF/anti-VEGF altered expression of genes encoding many members of the Wnt pathway. A subset of these genes was regulated in a way that is likely to contribute to control of the endothelial cell barrier. Namely, the VEGF-induced alteration of expression of such genes was reversed by anti-VEGF, and such adjustments occurred at times corresponding to changes in barrier function. While pharmacological and molecular approaches to activate the Wnt pathway had no effect on basal permeability, they suppressed VEGF-induced relaxation. Furthermore, anti-VEGF-mediated restoration of barrier function was unaffected by activation of the Wnt pathway.
Conclusions
VEGF/anti-VEGF engages multiple members of the Wnt pathway, and activating this pathway enforces the endothelial barrier by attenuating VEGF-induced relaxation. These data suggest that FDA-approved agents such as LiCl may be an adjuvant to anti-VEGF therapy for patients afflicted with blinding conditions including diabetic retinopathy.
Keywords: Wnt/beta-catenin signaling, VEGF, anti-VEGF, retina endothelial barrier, permeability
The fact that neutralizing vascular endothelial cell growth factor A (VEGF) within the eye is beneficial for many patients with blinding diseases such as diabetic macular edema (DME) and neovascular age-related macular degeneration (nAMD) demonstrates that VEGF is a clinically relevant regulator of vascular homeostasis within the retina. The encouraging results of ongoing clinical trials suggest that the Tie2 pathway is likely to also be a clinically relevant pathway.1,2 While anti-VEGF-directed therapies provide adequate benefit for many patients, alternate approaches are needed for a substantial proportion of individuals with nAMD and DME.
An approach to develop alternatives to anti-VEGFs is by considering the mechanism by which VEGF/anti-VEGF control the endothelial cell barrier. In addition to activating signaling events that modify components of cell-cell junctions,3–6 VEGF changes expression of thousands of genes, some of which encode known governors of blood vessel permeability.7 Such VEGF-regulated genes, which encode members of pathways that regulate vascular homeostasis, are likely to be involved with VEGF/anti-VEGF-mediated control of the endothelial cell barrier.
The Wnt/β-catenin pathway contributes to angiogenesis and barriogenesis during development,8–11 and recent advances have begun to reveal the underlying mechanisms. For instance, the classical Wnt pathway increases expression of Mfsd2a, and thereby suppresses caveolin1-dependent transcytosis across the blood-retinal barrier.12 Activation of the Wnt pathway also enforces the barrier function of cultured brain endothelial cells by reducing paracellular permeability.13 One of the Wnt ligand receptors (FZD7) restricts paracellular permeability and physically associates with components of the adherens junction complex.14 These mechanistic insights reinforce the concept that the Wnt pathway plays an important role in vascular homeostasis. The role of the Wnt pathway in VEGF/anti-VEGF-mediated control of the endothelial cell barrier has not been investigated.
We report that VEGF altered expression of many members of the Wnt pathway. Furthermore, activating the Wnt pathway enforced the barrier function of endothelial cells by reducing their responsiveness to VEGF. In contrast, activating the Wnt pathway had no effect on the efficacy of anti-VEGF.
Materials and Methods
Materials
Human retinal endothelial cells (HRECs) were purchased from Cell Systems (ACBRI 181; Kirkland, WA, USA). They were derived from donor A, a 26-year-old Caucasian male. Lonza endothelial cell basal medium-2 (EBM-2, CC3156) and Lonza SingleQuots endothelial cell growth medium-2MV (EGM- 2MV, CC4147) for tissue culture was procured from Lonza Bioscience (Verviers, Belgium). 293T cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The lentiviral plasmid, which was used to induce (TET-inducible) the expression of a constitutively active form of the β-catenin,15 was purchased from Addgene (114281, Cambridge, MA, USA). This β-catenin mutant is active because it lacks the first 45 amino acids, which include Ser 33, Ser 37, and Thr 41 that are phosphorylated by GSK3b to destabilize β-catenin.16 The lentiviral pXL010-Wnt dual (GFP-Fire) reporter, which has seven repeats of a TCF/LEF binding site and luciferase reporter,17 was purchased from Addgene (40588). QIAprep spin Miniprep Kit and QIAprep spin Maxiprep Kit were used to purify lentiviral plasmids from bacteria; they were purchased from QIAGEN (Hilden, Germany). The packaging plasmids pVSVg (Addgene 8454) and psPAX2 (Addgene 12260) were used to generate lentivirus. Lipofectamine used in transfection was purchased from Invitrogen (Waltham, MA, USA). Puromycin, which was used in drug selection, was purchased from Sigma-Aldrich (St. Louis, MO, USA). D-(+)-glucose (G7021), lithium chloride (7447-41-8), and doxycycline (D9891) were purchased from Sigma-Aldrich. Recombinant human IL-1β (200-01B) was purchased from PeproTech, Inc. (Rocky Hill, NJ, USA). Recombinant human VEGF-A (VEGF 165; 293-VE) and human tumor necrosis factor a (TNF-α) protein (210-TA) were purchased from R&D Systems (Minneapolis, MN, USA). Aflibercept (Eylea, Danvers, MA USA) was from Regeneron Pharmaceuticals, Inc. (Tarrytown, NY). Anti-β-catenin antibody (9562) was purchased from Cell Signaling Technology, Inc. (CST). Anti-β-actin antibody (ab8227) was purchased from Abcam (Boston, MA, USA). RNeasy Plus Mini Kit used for RNA isolation was obtained from QIAGEN (Hilden, Germany). The High-Capacity cDNA Reverse Transcription Kit used for cDNA synthesis and Fast SYBR Green Master Mix used for real-time PCR were purchased from Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA). Disposable electrode arrays (8W10E+ PC) used in transendothelial electrical resistance (TEER) assay for measuring cellular permeability were purchased from ECIS Cultureware (Applied BioPhysics, Inc., Troy, NY, USA). The luciferase assay system which was used to rapidly detect firefly luciferase activity was purchased from Promega (E1500, Madison, WI, USA).
Culture of Human Retinal Endothelial Cells and 293T Cells
To establish an in vitro model of DR, HRECs were cultured in complete Lonza medium (EBM-2 supplemented with the EGM-2MV SingleQuots kit) containing 30 mM D-glucose for at least 10 days; medium was changed every 24 hours. High glucose (HG)-treated HRECs were used for all experiments unless noted otherwise. 293T cells were cultured in DMEM medium containing 10% FBS and 1% P/S at 37°C, in a 5% CO2 cell incubator.
RNAseq
The goal of this series of experiments was to model VEGF- and anti-VEGF-mediated changes in gene expression in the context of diabetic retinopathy. To this end, triplicate dishes of confluent HG-treated HRECs were exposed to either VEGF vehicle, or VEGF (1 nM) for 48 hours and then harvested. Alternatively, cells were exposed to VEGF for 24 hours, then anti-VEGF for an additional 24 hours, and harvested. RNA was isolated and subjected to RNAseq analysis as previously described.7 Differentially expressed genes (DEGs) were identified by pair-wise comparison between experimental groups.
PCR Analysis
HG HRECs were seeded at 100% confluency. VEGF was added to a final concentration of 2 nM and then cultured for an additional 0.5, 1.5, or 8 hours. For the anti-VEGF group, after 8 hours of VEGF incubation, aflibercept (1 µM) was added for an additional eight hours. The cells were lysed, mRNA was isolated and used to synthesize cDNA. Quantitative PCR was performed using the real-time PCR HT7900 system (Applied Biosystems; Thermo Fisher Scientific). The threshold cycle (CT) of each transcript was normalized to the average CT of the housekeeping gene (β-actin). Fold differences were determined by the 2-∆∆CT method.
Western Blot Analysis
HG HRECs were treated with different agents (LiCl, VEGF, aflibercept, or doxycycline) for indicated time periods. After treatment, cells were then rinsed with ice-cold phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4) and lysed in electrophoresis 2X sample buffer (10 mM EDTA; 2% sodium dodecyl sulfate; 0.2 M 2-mercaptoethanol; 20% glycerol; 200 mM Tris-HCl, pH 6.8; and 0.2% bromophenol blue). Proteins were resolved on a 10% SDS-polyacrylamide gel and subjected to Western blot analysis.
TEER Measuring Cellular Permeability
Cell permeability was assessed by measuring changes in TEER using an electrical cell-substrate impedance sensing ZThera instrument (Applied Biophysics, Troy, NY, USA), which was housed in a standard tissue culture incubator that was maintained at 37°C and 5% CO2. Approximately 0.6 × 105 HG HRECs (1/8 of a confluent 10 cm dish) were resuspended in complete Lonza medium containing 30 mM D-glucose and seeded in each well of an 8-well chamber slide equipped with gold-coated microelectrodes (Applied Biophysics catalogue #8W10E+). The following day each well was inspected under the phase contrast microscope to verify that the monolayer was complete and overtly normal. At the start of the experiment impedance was typically 1500 ohms. Wells that had an impedance that was substantially different than this value were not used. While agents that influence barrier function (such as hydrocortisone)18–20 were present in the media, its concentration (200 nM) was not altered in any of the experiments.
After placing the chamber in the TEER instrument, the impedance was monitored until it stabilized (typically at least 30 minutes). The electric current passing through the endothelial monolayers was measured independently in each well. TEER was measured continuously and in real time before, during, and after the treatment of cells. When media were changed during a measurement period, the recording was paused until the media-change-induced noise subsided. When the Wnt activator (LiCl) was used, it was added 48 hours before addition of VEGF.
TEER data are presented as normalized impedance, which is the ratio of the impedance at a given time point/impedance at the starting point (time = 0). This ratio was calculated for each well. The starting impedance was typically very similar for all wells (variants were not included). Supplementary Figure S4 contains the same data set before and after normalization. Similarly, Supplementary Figures 2B, 3A, and 5A are presented as normalized impedance, and these data are presented without normalization in Supplementary Figure S5A, F, and H, respectively. Area under the curve (AUC) of unnormalized data sets was quantified using ImageJ.
Lentivirus Production
293T cells were transfected with the following three plasmids suspended in Lipofectamine: 15.5 µg transfer plasmids, (TET-inducible pCWXPGR-pTF-beta-catenin or pXL010-Wnt dual (GFP-Fire) reporter), 9 µg packaging plasmid (psPAX2), and 2.9 µg envelope plasmid (pVSVg). Three days postinfection the virus-containing medium was collected and stored at –80°C.
Infection of HRECs With Lentivirus
Passage 7 HG HRECs were cultured to approximately 90% confluence, whereupon virus-containing medium was added along with polybrene (final concentration of 4 µg/ml). After 24 hours the cells were cultured in a medium supplemented with 0.5 µg/mL puromycin. Medium was replaced with fresh medium every 2 days. The cells that proliferated in the selection medium were used for subsequent experiments.
Luciferase Assay
HG HREC-WNT reporter cells were seeded into 12-well plates. The cells were then treated with different agents (LiCl, VEGF, aflibercept, or doxycycline) for indicated time periods. After treatment, total cell lysates were extracted with 1X reporter lysis buffer (Promega) and 10 µg total cell proteins were used to determine luciferase activity by the Luciferase Assay System (Promega, Synergy H1, BioTek) using a manual luminometer (Berthold, Germany).
Statistical Analysis
The results are expressed as mean ± SD. Differences among groups were evaluated by analysis of variance; statistical significance of differences between groups was assessed using the t-test when indicated. Significance was defined as P < 0.05. Graphs were created using Microsoft Excel 2019 (Microsoft, Redmond, WA, USA).
Results
VEGF/Anti-VEGF Altered Expression of Many Wnt Pathway Genes
RNAseq analysis of HG-treated (to model diabetic retinopathy) primary HRECs that were stimulated with either VEGF, or VEGF and then anti-VEGF, revealed that many Wnt pathway genes were affected (Table 1). They responded in one of the following four ways. Expression of some genes was altered by VEGF, and such changes were reversed by anti-VEGF (e.g., DKK2). Other genes were regulated by VEGF, but not by anti-VEGF (e.g., FZD6). A way to think about such genes is that they constitute VEGF memory and are a potential explanation for why anti-VEGF is not beneficial for some patients. The third type of response that we observed was for genes whose expression changed in the same direction in response to both VEGF and anti-VEGF (e.g., CREBBP). Finally, there were genes that responded only to anti-VEGF (e.g., FZD2). Taken together these results indicate that many genes within the Wnt pathway are regulated by VEGF/anti-VEGF, and that there are at least four types of responses.
Table 1.
VEGF/Anti-VEGF Alters the Expression of Many Members of the Wnt Pathway
| Group | Statistically Significant Difference | No Statistically Significant Difference |
|---|---|---|
| Group 1: altered by VEGF; reversed by anti-VEGF | Control vs. VEGF VEGF vs. anti-VEGF | |
| Group 2: altered by VEGF; Not reversed by anti-VEGF | Control vs. VEGF | VEGF vs. anti-VEGF Control vs. anti-VEGF |
| Groups 3: altered by VEGF; altered by anti-VEGF in the same direction as VEGF | Control vs. VEGF VEGF vs. anti-VEGF | |
| Group 4: not altered by VEGF; altered by anti-VEGF | Control vs. anti-VEGF | Control vs. VEGF |
| The table indicates the types of comparisons that were the basis for the four groups. | ||
| Group 1 : Altered by VEGF; R eversed by A nti-VEGF | ||
| Gene Name | VEGF R esponse | Anti-VEGF R esponse |
| DKK1 | ↓ (P = 0.000) | ↑ (P = 0.000) |
| DKK2 | ↑ (P = 0.000) | ↓ (P = 0.000) |
| DKK3 | ↑ (P = 0.038) | ↓ (P = 0.020) |
| FZD1 | ↑ (P = 0.000) | ↓ (P = 0.004) |
| FZD8 | ↓ (P = 0.008) | ↑ (P = 0.000) |
| LRP5 | ↑ (P = 0.014) | ↓ (P = 0.017) |
| PSEN2 | ↑ (P = 0.000) | ↓ (P = 0.000) |
| CACYBP | ↓ (P = 0.045) | ↑ (P = 0.033) |
| CTNNB1 | ↑ (P = 0.000) | ↓ (P = 0.000) |
| JUN | ↓ (P = 0.000) | ↑ (P = 0.004) |
| FOSL1 | ↓ (P = 0.000) | ↑ (P = 0.007) |
| CCND2 | ↑ (P = 0.000) | ↓ (P = 0.001) |
| Group 2 : Altered by VEGF; N ot R eversed by A nti-VEGF | ||
| WNT10B | ↓ (P = 0.005) | |
| WNT2B | ↑ (P = 0.043) | |
| SFRP1 | ↓ (P = 0.011) | |
| FZD6 | ↑ (P = 0.002) | |
| CSNK1G2 | ↓ (P = 0.034) | |
| CSNK2A1 | ↓ (P = 0.003) | |
| DVL1 | ↓ (P = 0.001) | |
| AXIN1 | ↓ (P = 0.015) | |
| AXIN2 | ↑ (P = 0.022) | |
| PRKACA | ↓ (P = 0.023) | |
| PTPA | ↓ (P = 0.005) | |
| SKP1 | ↑ (P = 0.000) | |
| TBL1X | ↓ (P = 0.000) | |
| CTNNBIP1 | ↑ (P = 0.031) | |
| RBX1 | ↑ (P = 0.000) | |
| RUVBL1 | ↓ (P = 0.002) | |
| SMAD3 | ↓ (P = 0.000) | |
| CITED2 | ↑ (P = 0.028) | |
| TCF3 | ↓ (P = 0.003) | |
| TCF7 | ↓ (P = 0.016) | |
| TCF20 | ↓ (P = 0.020) | |
| TLE3 | ↓ (P = 0.013) | |
| MYC | ↓ (P = 0.001) | |
| Group 3 : Altered by VEGF; A ltered by A nti-VEGF in the S ame D irection as VEGF | ||
| CREBBP | ↓ (P = 0.029) | ↓ (P = 0.042) |
| CCND1 | ↓ (P = 0.000) | ↓ (P = 0.000) |
| TCF4 | ↑ (P = 0.002) | ↑ (P = 0.030) |
| Group 4 : N ot A ltered by VEGF; A ltered by A nti-VEGF | ||
| TLE2 | ↑ (P = 0.000) | |
| FZD2 | ↓ (P = 0.042) | |
| FZD4 | ↓ (P = 0.000) | |
| FZD5 | ↓ (P = 0.013) | |
| LRP6 | ↓ (P = 0.011) | |
Our previous RNAseq results showed that VEGF (1 nM) significantly altered (P < 0.05) expression of 4372 genes, and anti-VEGF (aflibercept, 500 nM) overcame the effect of VEGF for 279 of these genes in HG HRECs.7 Table 1 shows the subset of the data, which are members of Wnt pathway. The changes in expression between experimental groups is the basis for inclusion in a specific group.
The Wnt pathway regulates many facets of ocular vascular biology.21 Our goal was to investigate the role of the Wnt pathway in VEGF/anti-VEGF control of the endothelial cell barrier function (permeability). The expected behavior of Wnt pathway members that contributed to control of the endothelial cell barrier includes A) responsive to VEGF, B) such VEGF-induced changes are reversed by anti-VEGF, and C) this VEGF/anti-VEGF regulation of gene expression temporally aligns with VEGF/anti-VEGF-driven changes in barrier function.
Confluent HG HRECs establish a barrier, which can be relaxed with VEGF and then resealed by adding anti-VEGF (reference,7 Figs. 3, and 5). The time points at which gene expression was analyzed by qRT-PCR were chosen to align with changes in barrier function. At 0.5 and 1.5 hours the barrier was relaxing; at 8 hours it was maximally and stably relaxed; the barrier was reclosed within 8 hours after addition of anti-VEGF to such cells.7 Figure 1 shows the qRT-PCR results of genes whose expression was reproducibly altered by VEGF/anti-VEGF. Changes in expression occurred at different times post-VEGF, and thereby suggested that the Wnt pathway may contribute to multiple phases of barrier relaxation (initiation, execution, and persistence). RNAseq and qRT-PCR analysis indicated that VEGF-induced change in expression persisted to 24 or 48 hours if anti-VEGF was not added (reference7 and Supplementary Fig. S1). The nature of the change (direction and timing) for the genes shown in Figure 1 aligned with changes in barrier function and thereby supported the concept that the Wnt pathway contributes to VEGF/anti-VEGF control of the endothelial cell barrier.
Figure 3.
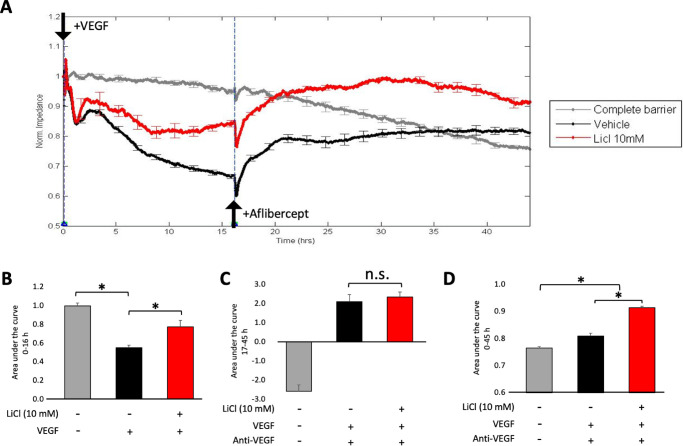
LiCl reduced VEGF's ability to relax the endothelial cell barrier. (A) Transendothelial electrical resistance (TEER) of HG HRECs treated in one of the following three ways. Cells were pretreated with vehicle (dH20) for 48 hours, VEGF (2 nM) was added, and after 16 hours anti-VEGF (1 µM) was added (black line; “Vehicle”). Cells were pretreated with LiCl (instead of dH20) for 48 hours, and LiCl (10 mM) was present together with VEGF and anti-VEGF (red line; “LiCl 10 mM”). No pretreatment, no VEGF or anti-VEGF (gray line; “Complete barrier”; gray line). Data were normalized to the initial TEER, which was measured at the 0 hour time point. Similar results were found in three independent experiments, which are shown as panels D–F of Supplementary Figure S5. Data are expressed as the mean ± SD for a single representative experiment. *P < 0.05. (B) The area under the curve for the first 16 hour course was quantified and normalized to the complete barrier group. *P < 0.05. (C) The area under the curve was quantified from 17 to 45 hours. n.s.: not significant. (D) The area under the curve was quantified for the entire time (0–45 hours). *P < 0.05.
Figure 5.
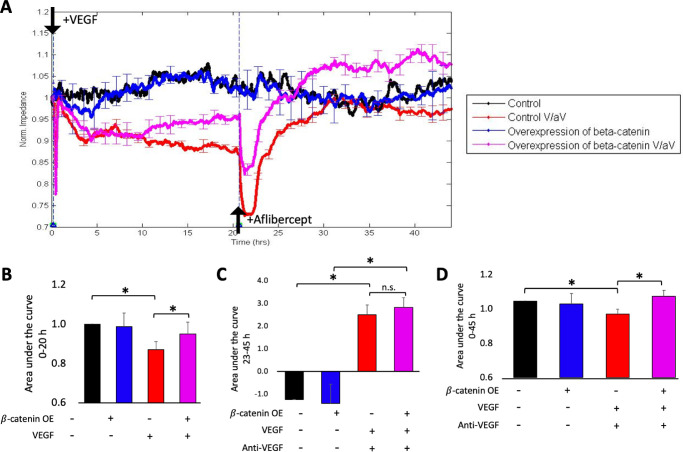
Overexpressing β-catenin reduced VEGF's ability to relax barrier. (A) Cells stably expressing the tet-regulated expression plasmid encoding a constitutively activated mutant of β-catenin (Addgene 114281) were pretreated with either vehicle (“Control”; black and red), or doxycycline (“Overexpression of β-catenin”; blue and pink) for 3 days and then barrier function was monitored following no further perturbation (black and blue) or in response to VEGF/anti-VEGF (pink and red). Data were normalized to the initial TEER, which was measured at the 0-hour time point. Data are expressed as the mean ± SD for a single representative experiment. *P < 0.05. Similar results were found in three independent experiments, which are shown as panels G–I of Supplementary Figure S5. (B) The area under the curve for the 0 to 20 hour period was quantified, and normalized to the “Control” or “Control V/aV” groups; V/aV indicates VEGF/anti-VEGF. *P < 0.05. (C) The area under the curve was quantified from 23 to 45 hours. n.s.: not significant. (D) The area under the curve was quantified for the entire time (0–45 hours). *P < 0.05.
Figure 1.
qRT-PCR analysis of the effect of VEGF/anti-VEGF on expression of Wnt pathway members. Triplicate dishes of HG HRECs were treated with either VEGF vehicle (black bar), or VEGF (1 nM; red bar) for the indicated duration and then harvested. The green bar indicates the response of cells that were treated with VEGF for 8 hours and then anti-VEGF for an additional 8 hours. Cells were lysed, RNA was extracted and subjected to qRT-PCR using specific primers. The level of expression in unstimulated cells (black bars) was set to 1.0. The data are expressed as the mean ± SD (n = 3–4). For VEGF-treated cells (red bars) the data are expressed as fold increase over unstimulated cells, *P < 0.05. The green bar shows the response of cells that were treated with VEGF for eight hours and then anti-VEGF for an additional 8 hours; statistically significant differences between these two experimental conditions are indicated by #P < 0.05. RNAseq and qRT-PCR analysis indicated that VEGF-induced change in expression persisted to 24 or 48 if anti-VEGF was not added (reference7 and Supplementary Fig. S1). Similar results we observed in three independent experiments.
The Wnt Pathway Activator (LiCl) Perturbed VEGF's Ability to Relax the Barrier
All 10 Wnt pathway family members, which were regulated in ways that were likely to contribute to VEGF/anti-VEGF control of the barrier (see above), have been reported to either activate or suppress the Wnt pathway (Table 2). These previous reports do not unanimously indicate if VEGF activates or inhibits the Wnt pathway. For instance, while VEGF increased expression of DKK2, CTNNB1, APC, and CSNK1A1, APC and CSNK1A1 are reported to inhibit the Wnt pathway, CTNNB1 activates it, while DKK2 either inhibits or activates it (Table 2). Thus, the work of other investigators, which was done in a variety of experimental systems and cell types, does not point to Wnt pathway members that are likely to be contributing, or even predict whether VEGF-induced relaxation of the barrier involves activation or inhibition of the Wnt pathway.
Table 2.
Previously Reported Effects of VEGF on Expression of Members of the Wnt pathway and How Such Changes Influence the Activity of the Pathway
| Gene | Response to VEGF | Effect on Wnt/β-Catenin Pathway |
|---|---|---|
| DKK2 | Upregulation | Inhibition26–29 or activation30 |
| CTNNB1 | Upregulation | Activation31 |
| APC | Upregulation | Inhibition32,33 |
| CSNK1A1 | Upregulation | Inhibition34,35 |
| SFRP4 | Downregulation | Inhibition36,37 |
| FZD1 | Upregulation | Activation38–40 |
| SOX17 | Upregulation | Inhibition41–43 or activation44 |
| JUN | Downregulation | Inhibition45 or activation46 |
| SFRP1 | Downregulation | Inhibition47–49 or activation50 |
| WNT4 | Upregulation | Inhibition51 or activation52 |
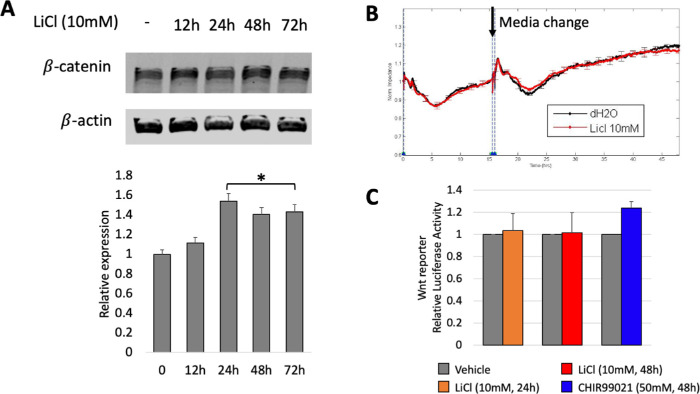
Our first approach to determine the effect of activating the Wnt pathway on basal and VEGF/anti-VEGF-dependent control of the barrier was to treat cells with LiCl, which stabilizes β-catenin and thereby activates the canonical Wnt pathway. Dose-response experiments indicated that up to 10 mM had no effect on either basal permeability or viability, whereas at concentrations of 20 mM and above, the cells perished and consequently barrier function declined precipitously. Time-course experiments indicated that 10 mM LiCl for 24 to 72 hours modestly increased the level of β-catenin protein, without perturbing the ability of cells to establish a barrier under basal (unchallenged) conditions (Figs. 2A and B). The magnitude of the increase in β-catenin protein was insufficient to alter β-catenin-dependent transcriptional activity (Fig. 2C), which was assessed in HG HRECs stably expressing the TCF/LEF reporter. Expression of negative regulators of β-catenin-dependent transcription such as CTNNBIP1 and CITED2 may suppress β-catenin transcriptional activity in LiCl2-treated cells.
Figure 2.
LiCl modestly elevated the level of β-catenin and had no effect on basal permeability or β-catenin-dependent transcriptional activity. (A) HG HRECs were treated with 10 mM LiCl for the indicated duration, then lysed and the clarified cell lysates were subjected to Western blot analysis. The level of β-catenin expression was quantified and normalized to the level of β-actin. The bar graph was generated from three independent repeats. Data are expressed as means ± SD. There was a statistically significant increase in the level of β-catenin at the 24-, 48-, and 72-hour time points. (B) The transendothelial electrical resistance (TEER) of confluent monolayers of HG HRECs was measured by electrical cell-substrate impedance sensing (ECIS) for 48 hours following the addition of either LiCl vehicle (dH2O; black) or LiCl (10 mM; red). At the 16-hour time point, cells received a media change and dH2O and LiCl were added again. A high TEER value indicates effective barrier function, that is, low permeability. The data are expressed as the mean ± SD (n = 3–4). Similar results were found in three independent experiments, which are shown as panels A–C of Supplementary Figure S5. (C) HG HRECs stably expressing a TCF/LEF driven reporter plasmid were treated with either vehicle (dH2O), LiCl (10 mM) for 24 or 48 hours, or CHIR99021 (50 nM) for 48 hours, lysed, and luciferase activity was quantified as described in the Materials and Methods section. Luciferase activity was normalized to the internal transfection control, and values for vehicle controls were set as 1-fold induction. The data presented are mean ± SD of at least three independent experiments performed in triplicates.
We also observed that LiCl-treated cells were less responsive to VEGF. While there was no effect on acute relaxation, which occurs within minutes after adding VEGF, subsequent relaxation was attenuated in LiCl treated cells (Figs. 3A and B). Figure 3B quantifies the data set during the VEGF-induced relaxation phase of the experiment (0–16 hours) and demonstrates that there was a statistically significant difference between the extent of VEGF-induced relaxation in vehicle versus LiCl-treated cells. Both VEGF-induced relaxation and the partial suppression by LiCl persisted for up to 45 hours (Supplementary Fig. S2).
In contrast, LiCl did not alter the cells’ response to anti-VEGF. After relaxing the barrier with VEGF, we added anti-VEGF and continued to monitor permeability until the 45-hour time point. As previously reported, anti-VEGF at first further relaxed the barrier before reclosing it,7 which is what was observed with the “Vehicle” (Fig. 3A and C) cells. The effectiveness of anti-VEGF was comparable in vehicle and LiCl-treated cells (Fig. 3A and C). Panel D shows the area under the curve for the entire experiment (0–45 hours). We conclude that LiCl attenuated VEGF-mediated permeability and had no effect on the ability of cells to respond to anti-VEGF.
In contrast to the attenuating effect of LiCl on VEGF-induced barrier relaxation, LiCl had either no effect (IL1-β), or slightly promoted (TNF-α) cytokine-induced permeability (Supplementary Fig. S3). These results indicate that the impact of LiCl on barrier function was not uniform for all inducers of permeability.
Overexpressing β-Catenin Attenuated VEGF Induced Relaxation
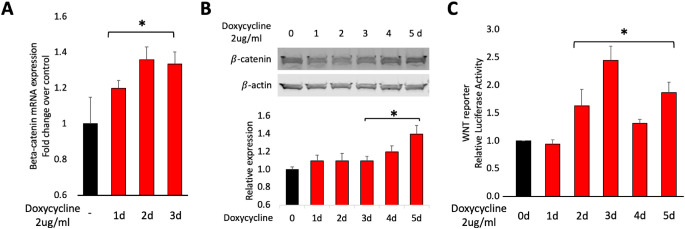
To complement the pharmacological approach (Figs. 2 and 3), we tested the effect of overexpressing β-catenin on VEGF/anti-VEGF control of the barrier. To this end, we generated HG HRECs that stably expressed a doxycycline-regulated, constitutively active β-catenin mutant cDNA.15 Addition of doxycycline first increased the level of β-catenin mRNA, and then elevated the level of β-catenin protein (Figs. 4A and B). β-catenin-dependent transcriptional activity was also increased (Fig. 4C), even though the level of β-catenin overexpression was comparable to what occurred in response to LiCl (Figs. 2 and 4). A likely explanation for this difference is that the molecular approach involved a constitutively activated mutant of β-catenin, whereas the pharmacological approach elevated the level of endogenous, wild type β-catenin.
Figure 4.
Characterization of β-catenin overexpressing cells. In this series of experiments, we used cells stably expressing a tet-regulated expression plasmid encoding a constitutively activated mutant of β-catenin (Addgene 114281). (A) Cells were treated with doxycycline (2 µg/mL) for 0, 1, 2, or 3 days. RNA was extracted and subjected to qRT-PCR using primers specific for either β-catenin or β-actin. The expression of β-catenin was quantified and normalized to the level of β-actin. The red bars display the doxycycline-induced fold change over untreated cells (black bar); *P < 0.05. The data are expressed as the mean ± SD (n = 3–4). Similar results we observed in three independent experiments. (B) Cells were treated with doxycycline (2 µg/mL) for the indicated duration, then lysed, and the clarified cell lysates were subjected to Western blot analysis. The level of β-catenin expression was quantified and normalized to the level of β-actin. The bar graph was generated from three independent repeats. Data are expressed as means ± SD. There was a statistically significant increase in the level of β-catenin at 3, 4, and 5 days. (C) Cells co-expressing the β-catenin expression vector and TCF/LEF reporter were treated with either vehicle (dH2O) or LiCl (10 mM) for the indicated duration, lysed, and luciferase activity was quantified as described in the Materials and Methods section. Luciferase activity was normalized to the internal transfection control, and values for vehicle controls were set as 1-fold induction. The data presented are mean ± SD of at least three independent experiments performed in triplicates. There was a statistically significant increase luciferase activity at 2, 3, 4, and 5 days.
While the basal barrier function was unaffected by β-catenin overexpression (Supplementary Fig. S4), VEGF-driven barrier relaxation was attenuated in the overexpressing cells (Figs. 5A and B). Furthermore, overexpression of activated β-catenin had no effect on the ability of anti-VEGF to reclose the barrier (Figs. 5A and C). Panel D shows the area under the curve for the entire experiment (0–45 hours). These results are consistent with the pharmacological approach, which also indicated that activation of the Wnt pathway suppresses VEGF-induced barrier relaxation and did not influence the ability of cells to respond to anti-VEGF.
Discussion
We report that VEGF/anti-VEGF engages many members of the Wnt pathway. Furthermore, activating the Wnt pathway reduced VEGF-driven relaxation of the endothelial cell barrier, and had no effect on the ability of anti-VEGF to reclose the barrier. These observations suggest that approaches to activate the Wnt pathway may be beneficial for patients afflicted by VEGF-driven disease.
Expression pattern is one way to identify Wnt family members that govern the endothelial cell barrier. For instance, those genes whose expression is altered by VEGF, and reversed by anti-VEGF are most likely to be regulators of the barrier (first section of Table 1). Members of the Wnt pathway whose expression is not regulated in this manner are probably not participants in this context and/or their input occurs at times that we did not consider.
Our RNAseq results indicate that VEGF increased expression of CTNNB1 and anti-VEGF reduced it (Table 1). These observations predict that pharmacologically or molecularly stabilizing β-catenin would relax the endothelial barrier. Surprisingly, such interventions had no effect on basal permeability (Fig. 2B and Figs. 5A, B), rather they enforced the barrier when cells were exposed to VEGF (Figs. 3 and 5). Table 1 demonstrates that there are many additional Wnt pathway family members that were regulated by VEGF. We conclude that while VEGF/anti-VEGF-dependent regulation of the endothelial barrier involves the Wnt pathway, further investigation is necessary to understand how the participating family members contribute.
The existence of genes that are responsive to VEGF, but unresponsive to anti-VEGF (second section of Table 1) suggests the concept of VEGF memory; that the molecular profile of cells remains altered even after the VEGF responsible for such changes has been neutralized. VEGF memory is in some ways comparable to metabolic memory, which involves hyperglycemia-driven epigenetic modifications that persist even when glycemia is lowered.22,23 VEGF memory is a novel explanation for the nonuniform response of patients to anti-VEGF. Traditional explanations include A) that the level of VEGF exceeds the neutralizing capacity of anti-VEGFs and B) that non-VEGFs (such as cytokines) are drivers of pathology. Table 1 lists 23 Wnt pathway members that are VEGF memory genes, that is, anti-VEGF is unable to overcome the change in expression induced by VEGF. Such VEGF-induced changes persist even after VEGF has been neutralized. Approaches that target this subset of the Wnt pathway may provide benefit to those patients that respond suboptimally to anti-VEGF. As the details of VEGF memory emerge, it may become possible to erase VEGF memory using Wnt-targeted therapies such as LiCl.
Activating the Wnt pathway is not the only effect of LiCl. LiCl inhibits the activity of IMPase (inositol monophosphatase), an enzyme that contributes to regeneration of PI/PIP2 (phosphatidylinositol/phosphatidylinositol bisphosphate).24 Several of the signaling enzymes that are activated by VEGF (phospholipase C gamma and phosphoinositide 3 kinase) compete for PIP2.24,25 By impairing regeneration of PIP2, LiCl may perturb the signaling events downstream of the activated VEGF receptor. An alternative approach to increase the level of β-catenin (overexpressing a constitutively activated β-catenin c-DNA) had a similar effect as treating cells with LiCl. Taken together, it appears that activating the Wnt pathway was the reason that LiCl enforced barrier function in this VEGF-based context. Regardless of the mechanism of action, these observations beg the question of whether LiCl, a relatively safe, FDA-approved agent, could be used as an adjuvant to anti-VEGF.
While both approaches to elevate β-catenin resulted in a comparable elevation in the level of β-catenin proteins, only the molecular approach resulted in an increase in β-catenin-dependent transcriptional activity (Figs. 2 and 4). A likely explanation is that LiCl stabilized the endogenous, wild type β-catenin, whereas cDNA that was introduced into cells coded for a constitutively activated version of β-catenin.
Having discovered that VEGF/anti-VEGF engage the Wnt pathway to regulate barrier function, the next logical step is to identify the genes that are responsible. Table 1 lists the candidates that will be evaluated in future studies.
Supplementary Material
Acknowledgments
Supported by grants for the Juvenile Diabetes Research Foundation, Illinois Society to Prevent Blindness, National Institute of Health (EY031350) and an unrestricted grant from the Research to Prevent Blindness Foundation. The pCWXPGR-pTF-beta-catenin was a gift from Patrick Salmon (Addgene plasmid #114281; http://n2t.net/addgene:114281; RRID: Addgene_114281).
Disclosure: Y. Li, None; B. Baccouche, None; O. Olayinka, None; A. Serikbaeva, None; A. Kazlauskas, None
References
- 1.Chakravarthy U, Bailey C, Brown D, et al.. Phase I trial of anti-vascular endothelial growth factor/anti-angiopoietin 2 bispecific antibody RG7716 for neovascular age-related macular degeneration. Ophthalmol Retina. 2017; 1: 474–485. [DOI] [PubMed] [Google Scholar]
- 2.Campochiaro PA, Khanani A, Singer M, et al.. Enhanced benefit in diabetic macular edema from AKB-9778 Tie2 activation combined with vascular endothelial growth factor suppression. Ophthalmology. 2016; 123: 1722–1730. [DOI] [PubMed] [Google Scholar]
- 3.Claesson-Welsh L.Vascular permeability–the essentials. Ups J Med Sci. 2015; 120: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejana E.Endothelial adherens junctions: implications in the control of vascular permeability and angiogenesis. J Clin Invest. 1997; 100: S7–10. [PubMed] [Google Scholar]
- 5.Komarova YA, Kruse K, Mehta D, Malik AB.. Protein interactions at endothelial junctions and signaling mechanisms regulating endothelial permeability. Circ Res. 2017; 120: 179–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goddard LM, Iruela-Arispe ML.. Cellular and molecular regulation of vascular permeability. Thromb Haemost. 2013; 109: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Yan Z, Chaudhry K, Kazlauskas A.. The renin-angiotensin-aldosterone system (RAAS) is one of the effectors by which vascular endothelial growth factor (VEGF)/anti-VEGF controls the endothelial cell barrier. Am J Pathol. 2020; 190: 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, Barres BA.. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci USA. 2009; 106: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebner S, Corada M, Bangsow T, et al.. Wnt/beta-catenin signaling controls development of the blood-brain barrier. J Cell Biol. 2008; 183: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, McMahon AP.. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008; 322: 1247–1250. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Wang Y, Tischfield M, et al.. Canonical WNT signaling components in vascular development and barrier formation. J Clin Invest. 2014; 124: 3825–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Liu CH, Huang S, et al.. Wnt signaling activates MFSD2A to suppress vascular endothelial transcytosis and maintain blood-retinal barrier. Sci Adv. 2020; 6: eaba7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laksitorini MD, Yathindranath V, Xiong W, Hombach-Klonisch S, Miller DW.. Modulation of Wnt/beta-catenin signaling promotes blood-brain barrier phenotype in cultured brain endothelial cells. Sci Rep. 2019; 9: 19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira Tojais N, Peghaire C, Franzl N, et al.. Frizzled7 controls vascular permeability through the Wnt-canonical pathway and cross-talk with endothelial cell junction complexes. Cardiovasc Res. 2014; 103: 291–303. [DOI] [PubMed] [Google Scholar]
- 15.Boitard M, Bocchi R, Egervari K, et al.. Wnt signaling regulates multipolar-to-bipolar transition of migrating neurons in the cerebral cortex. Cell Rep. 2015; 10: 1349–1361. [DOI] [PubMed] [Google Scholar]
- 16.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT.. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996; 10: 1443–1454. [DOI] [PubMed] [Google Scholar]
- 17.Azarin SM, Lian X, Larson EA, Popelka HM, de Pablo JJ, Palecek SP.. Modulation of Wnt/beta-catenin signaling in human embryonic stem cells using a 3-D microwell array. Biomaterials. 2012; 33: 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos CJ, Lin C, Liu X, Antonetti DA.. The EPAC-Rap1 pathway prevents and reverses cytokine-induced retinal vascular permeability. J Biol Chem. 2018; 293: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonetti DA, Wolpert EB, DeMaio L, Harhaj NS, Scaduto RC Jr. Hydrocortisone decreases retinal endothelial cell water and solute flux coincident with increased content and decreased phosphorylation of occludin. J Neurochem. 2002; 80: 667–677. [DOI] [PubMed] [Google Scholar]
- 20.Harhaj NS, Felinski EA, Wolpert EB, Sundstrom JM, Gardner TW, Antonetti DA.. VEGF activation of protein kinase C stimulates occludin phosphorylation and contributes to endothelial permeability. Invest Ophthalmol Vis Sci. 2006; 47: 5106–5115. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Liu CH, Huang S, Chen J.. Wnt signaling in vascular eye diseases. Prog Retin Eye Res. 2019; 70: 110–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Miao F, Paterson AD, et al.. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci USA. 2016; 113: E3002–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper ME, El-Osta A.. Epigenetics: mechanisms and implications for diabetic complications. Circ Res. 2010; 107: 1403–1413. [DOI] [PubMed] [Google Scholar]
- 24.Stratman AN, Farrelly OM, Mikelis CM, et al.. Anti-angiogenic effects of VEGF stimulation on endothelium deficient in phosphoinositide recycling. Nat Commun. 2020; 11: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Im E, Kazlauskas A.. Regulating angiogenesis at the level of PtdIns-4,5-P(2). EMBO J. 2006; 25: 2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niehrs C.Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006; 25: 7469–7481. [DOI] [PubMed] [Google Scholar]
- 27.Baetta R, Banfi C. Dkk (Dickkopf) Proteins. Arterioscler Thromb Vasc Biol. 2019; 39: 1330–1342. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Zhang S, Gu L, Di W.. Epigenetic silencing of DKK2 and Wnt signal pathway components in human ovarian carcinoma. Carcinogenesis. 2012; 33: 2334–2343. [DOI] [PubMed] [Google Scholar]
- 29.Xu W, Pang K, Zhou ZG, et al.. Dickkopf 2 promotes proliferation and invasion via Wnt signaling in prostate cancer. Mol Med Rep. 2016; 14: 2283–2288. [DOI] [PubMed] [Google Scholar]
- 30.Devotta A, Hong CS, Saint-Jeannet JP.. Dkk2 promotes neural crest specification by activating Wnt/beta-catenin signaling in a GSK3beta independent manner. Elife. 2018; 7, doi: 10.7554/eLife.34404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald BT, Tamai K, He X.. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009; 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker TW, Neufeld KL.. APC controls Wnt-induced beta-catenin destruction complex recruitment in human colonocytes. Sci Rep. 2020; 10: 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sierra J, Yoshida T, Joazeiro CA, Jones KA.. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006; 20: 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Li Y, Semenov M, et al.. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002; 108: 837–847. [DOI] [PubMed] [Google Scholar]
- 35.Hammerlein A, Weiske J, Huber O.. A second protein kinase CK1-mediated step negatively regulates Wnt signalling by disrupting the lymphocyte enhancer factor-1/beta-catenin complex. Cell Mol Life Sci. 2005; 62: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K, Kitazawa R, Kondo T, et al.. Diabetic osteopenia by decreased beta-catenin signaling is partly induced by epigenetic derepression of sFRP-4 gene. PLoS One. 2014; 9: e102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakanishi R, Akiyama H, Kimura H, et al.. Osteoblast-targeted expression of Sfrp4 in mice results in low bone mass. J Bone Miner Res. 2008; 23: 271–277. [DOI] [PubMed] [Google Scholar]
- 38.L'Episcopo F, Serapide MF, Tirolo C, et al.. A Wnt1 regulated Frizzled-1/beta-catenin signaling pathway as a candidate regulatory circuit controlling mesencephalic dopaminergic neuron-astrocyte crosstalk: therapeutical relevance for neuron survival and neuroprotection. Mol Neurodegener. 2011; 6: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolben T, Perobner I, Fernsebner K, et al.. Dissecting the impact of Frizzled receptors in Wnt/beta-catenin signaling of human mesenchymal stem cells. Biol Chem. 2012; 393: 1433–1447. [DOI] [PubMed] [Google Scholar]
- 40.Planutis K, Planutiene M, Nguyen AV, Moyer MP, Holcombe RF.. Invasive colon cancer, but not non-invasive adenomas induce a gradient effect of Wnt pathway receptor frizzled 1 (Fz1) expression in the tumor microenvironment. J Transl Med. 2013; 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinner D, Kordich JJ, Spence JR, et al.. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007; 27: 7802–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Yang WT, Zheng PS, Liu XF.. SOX17 restrains proliferation and tumor formation by down-regulating activity of the Wnt/beta-catenin signaling pathway via trans-suppressing beta-catenin in cervical cancer. Cell Death Dis. 2018; 9: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M.. SOX17 antagonizes WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010; 5: 743–749. [DOI] [PubMed] [Google Scholar]
- 44.Mukherjee S, Chaturvedi P, Rankin SA, et al.. Sox17 and beta-catenin co-occupy Wnt-responsive enhancers to govern the endoderm gene regulatory network. Elife. 2020; 9, doi: 10.7554/eLife.58029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu D, Fang W, Han A, et al.. c-Jun N-terminal kinase 1 interacts with and negatively regulates Wnt/beta-catenin signaling through GSK3beta pathway. Carcinogenesis. 2008; 29: 2317–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP, Li L.. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol. 2008; 180: 1087–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J.. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008; 121: 737–746. [DOI] [PubMed] [Google Scholar]
- 48.Finch PW, He X, Kelley MJ, et al.. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA. 1997; 94: 6770–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawano Y, Kypta R.. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003; 116: 2627–2634. [DOI] [PubMed] [Google Scholar]
- 50.Zhong X, Desilva T, Lin L, et al.. Regulation of secreted Frizzled-related protein-1 by heparin. J Biol Chem. 2007; 282: 20523–20533. [DOI] [PubMed] [Google Scholar]
- 51.Bernard P, Fleming A, Lacombe A, Harley VR, Vilain E.. Wnt4 inhibits beta-catenin/TCF signalling by redirecting beta-catenin to the cell membrane. Biol Cell. 2008; 100: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyons JP, Mueller UW, Ji H, et al.. Wnt-4 activates the canonical beta-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/beta-catenin activity in kidney epithelial cells. Exp Cell Res. 2004; 298: 369–387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.