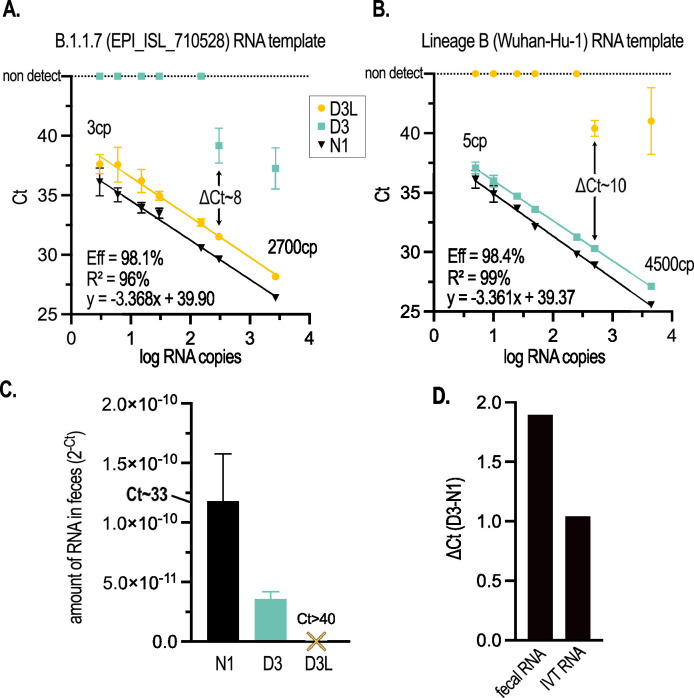
Fig. 2.
The D3L and D3 qRT-PCR assays are highly sensitive and sufficiently discriminate targeted alleles on IVT RNA templates and in COVID-19 patient fecal samples. (A) Standard curve of N1, D3L, and D3 assays performed with serial dilutions of a synthetic B.1.1.7 genomic RNA template (GISAID accession ID EPI_ISL_710528, Twist Biosciences). (B) Standard curve of N1, D3L, and D3 assays performed with serial dilutions of a synthetic non-B1.1.7 genomic RNA template (Wuhan-Hu-1 isolate). For both standard curves, N1 RNA copy number (cp) determined by RT-ddPCR is indicated. In panels (A) and (B), error bars represent the standard deviation of the mean of 4 qRT-PCR replicates. Calculated primer efficiencies ([10-1/m -1]*100), linear fit, and equation of the line of regression are indicated for D3L (A) and D3 (B). (C) The B.1.1.7 allele (D3L) is not detectable in RNA extracted from the pooled feces of 2 human COVID-19 patients admitted to an Ottawa hospital prior to the emergence of B.1.1.7 in Canada; while both N1 and D3 (non-B.1.1.7) alleles are detected. Error bars represent the standard deviation of the mean of 2-3 qRT-PCR replicates. (D) D3 alleles amplify approx. 1 Ct later than the N1 allele on IVT RNA, with data derived from (B); and approx. 2 Cts later in patient feces, with data derived from (C).