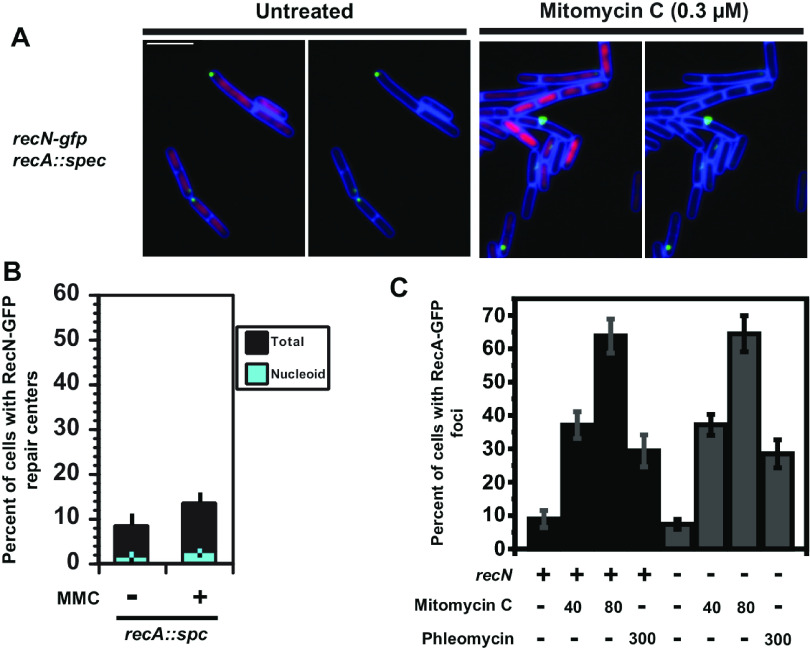
FIG 5.
RecA is required for the formation of RecN-GFP foci on the nucleoid in response to DNA damage. (A) Representative micrographs of B. subtilis cells with RecN-GFP and in cells deficient for recA left untreated or challenged with mitomycin C. Cell membranes were stained with FM 4-64 (pseudocolored blue), and the nucleoid was imaged with DAPI (pseudocolored magenta). (B) Bar graph shows the percentage of cells with foci that were nucleoid associated (cyan) or off the nucleoid (black) in the presence or absence of DNA damage in cells deficient for recA. Error bars represent the 95% confidence interval. For the untreated condition, n = 1,008, and for MMC treatment, n = 1,336 for cells scored. (C) Shown is a graph with the percentage of cells with RecA-GFP foci challenged with mitomycin C or phleomycin that were wild type for recN or carrying the recN::cm allele. The number of cells scored for each condition are the following: for recA-GFP and recN::cm untreated, n = 1,211; for 40 nM MMC, n = 924; for 80 nM MMC, n = 301; for 300 nM phleomycin, n = 446; for recA-GFP and wild-type recN, untreated n = 479; 40 nM MMC, n = 547; for 80 nM MMC, n = 340; for 300 nM phleomycin, n = 347. Error bars represent the 95% confidence intervals.