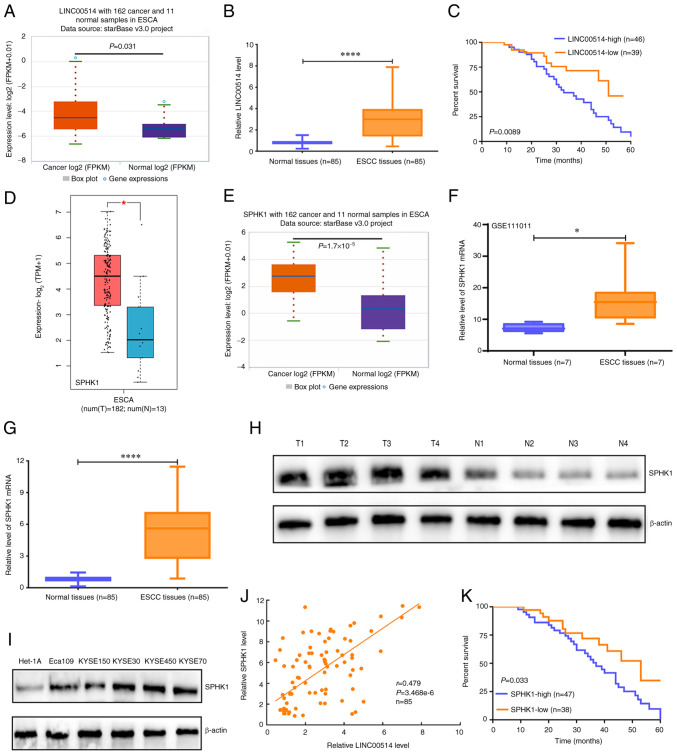
Figure 1.
Expression and prognostic value of LINC00514 and SPHK1 in ESCC. (A) starBase was used to investigate the expression of LINC00514 in 162 esophageal cancer samples and 11 normal samples. (B) Reverse transcription-quantitative PCR was used to evaluate the expression levels of LINC00514 in 85 ESCC and paired normal samples using a Wilcoxon signed rank test. ****P<0.0001. (C) A log-rank test was used to analyze the effects of LINC00514 expression on the survival rate of patients with ESCC. (D) Gene Expression Profiling Interactive Analysis online software analysis of SPHK1 level in 182 ESCA and 13 normal samples. (E) starBase analysis was performed to determine the expression of SPHK1 in 162 ESCA and 11 normal samples. (F) The GSE111011 Gene Expression Omnibus dataset was used to detect the levels of SPHK1 in seven ESCC and paired normal samples using paired a Student's t-test. *P<0.05. (G) Reverse transcription-quantitative PCR assay was used to determine the SPHK1 levels in 85 ESCC and paired normal samples, and the data was analyzed using a Wilcoxon signed rank test. (H) SPHK1 protein expression in four randomly selected paired tissues. β-actin was used as the loading control. (I) SPHK1 protein expression in ESCC cell lines (Eca109, KYSE150, KYSE30, KYSE450 and KYSE70) and the normal esophageal epithelial cell line Het-1A. β-actin was used as a loading control. (J) Spearman's rank correlation analysis of the correlation between LINC00514 and SPHK1 expression in the 85 ESCC and paired normal samples. (K) Log-rank analysis of the effect of SPHK1 expression on the survival rate of patients with ESCC. ****P<0.0001. LINC00514, long intergenic nonprotein-coding RNA 00514; ESCC, esophageal squamous cell carcinoma; SPHK1, sphingosine kinase 1; ESCA, esophageal cancer; T, tumor; N, normal.