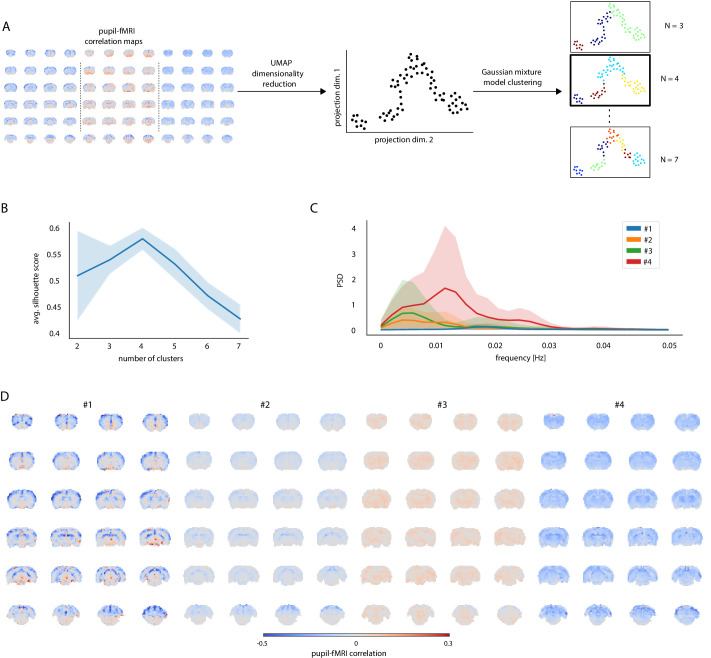
Figure 2. Clustering of trials with distinct pupil–fMRI correlation patterns.
(A) Schematic of the clustering procedure. UMAP is used to reduce the dimensionality of all individual-trial correlation maps to 72 dimensions. A 2D UMAP-projection of the real data is shown. Each dot represents a single trial. The trials are clustered using Gaussian mixture model clustering. Different numbers of clusters are evaluated. (B) The final number of clusters is selected based on silhouette analysis. The highest average silhouette score is obtained with k = 4 clusters. Shaded area shows standard deviations. (C) Pupil power spectral density estimates (PSD) of each of the four clusters. Signals were downsampled to match the fMRI sampling rate. Shaded areas show standard deviations. (D) Cluster-specific correlation maps based on concatenated signals belonging to the respective groups.
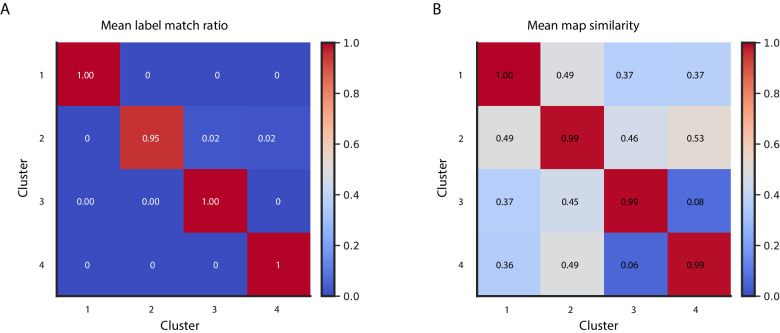
Figure 2—figure supplement 1. Cluster reproducibility across 100 repetitions with random UMAP and GMM initializations.
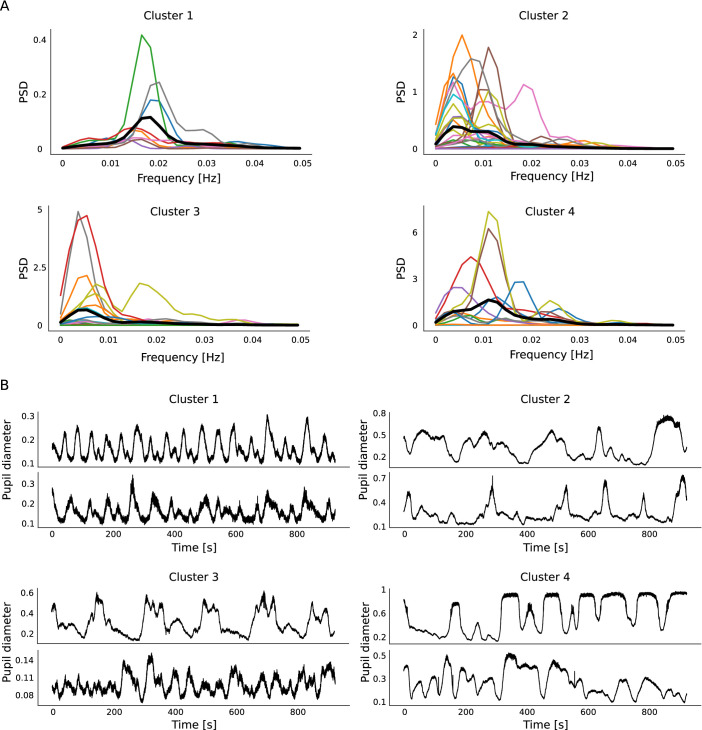
Figure 2—figure supplement 2. Cluster-specific pupil fluctuation features.
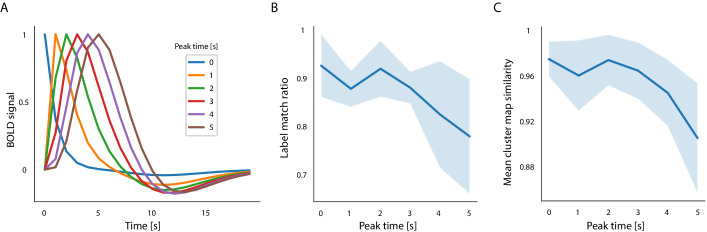
Figure 2—figure supplement 3. Clustering reproducibility across 100 clustering repetitions based on HRF-convolved pupil signals.
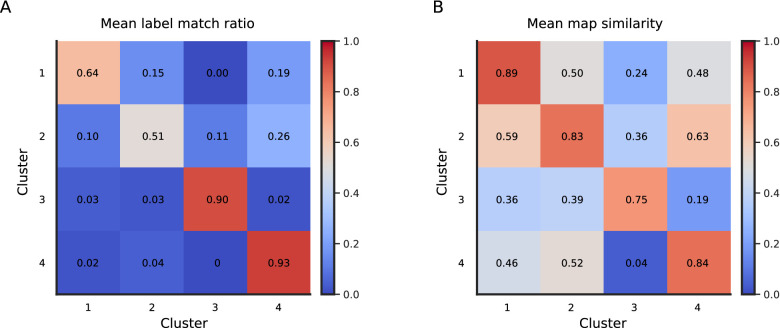
Figure 2—figure supplement 4. Cluster reproducibility across 100 repetitions of split-halves clustering.
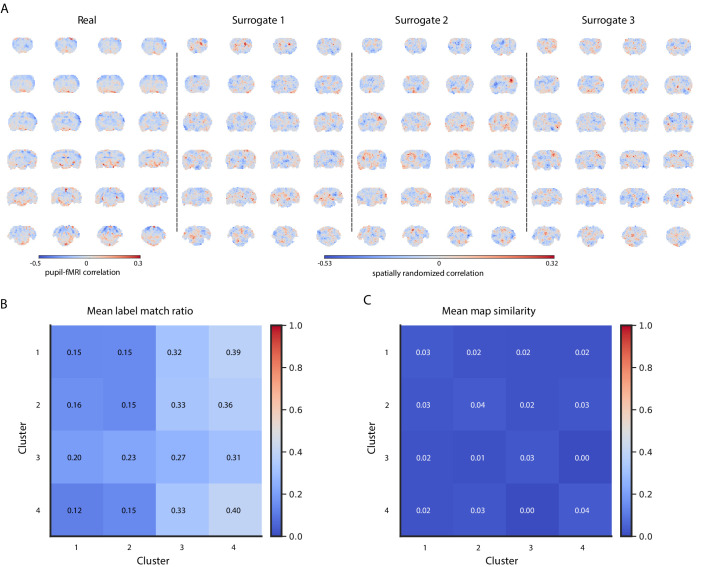
Figure 2—figure supplement 5. Cluster reproducibility across 100 sets of artificially generated surrogates with values and spatial autocorrelations matching those of real maps.
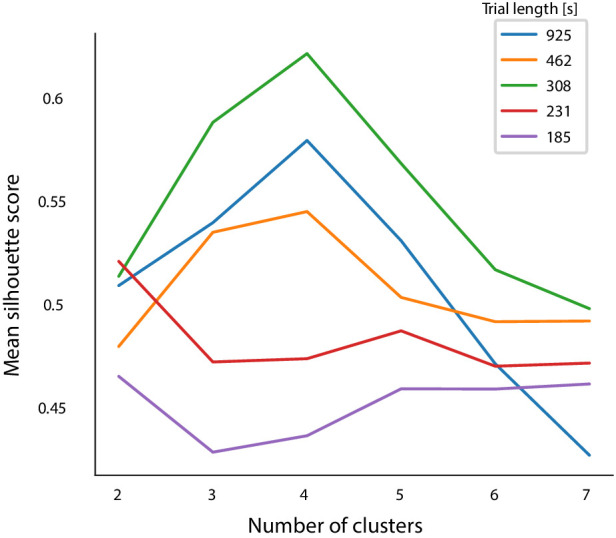
Figure 2—figure supplement 6. Mean silhouette scores based on 100 clustering repetitions performed on shorter trials.