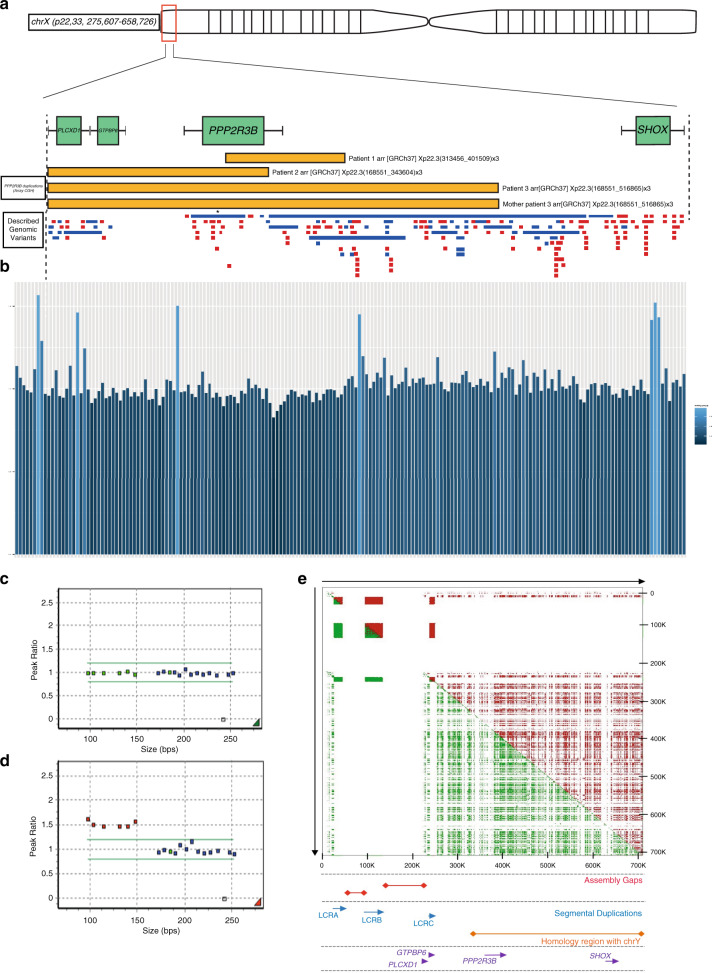
Fig. 1. Germline duplications involving PPP2R3B are found at increased frequency in individuals with melanocytic neoplasia.
(a) Schematic of Xp22.33 demonstrating the location of three novel duplications (yellow) found in 24 congenital melanocytic nevi (CMN) patients using whole-genome array comparative genomic hybridization (CGH) of leukocyte DNA, with one identical parental duplication demonstrating inheritance. Previously described copy-number variants in that region are shown below, duplications in blue, deletions in red, with each bar representing a single publication. The publication representing a duplication involving PPP2R3B described a single variant in a cohort of approximately 36,000 (asterisk; see text for details), confirming that the CMN duplications are rare in the normal population. (b) PPP2R3B duplications in a UK nonsyndromic melanoma cohort (4 duplications in nonselected cohort n = 168), and CMN cohort (3 duplications in known preselected cohort n = 5) shown by targeted next-generation sequencing (NGS) of PPP2R3B, in addition to the two telomeric genes (GTPBP6 and PLCXD1) and the next centromeric gene (SHOX). Data represent the ratio of corrected read depth (see text for details) across the whole of PPP2R3B with respect to the ratio across the whole of SHOX. Each bar represents an individual patient. PPP2R3B duplications called are shown in light blue: validation of the array CGH findings in the three CMN patients are clustered to the right of the figure, and new duplications in the melanoma cohort in the rest of the figure (n = 4, 2.4%). Validation of PPP2R3B duplications detected by array CGH. Custom-designed multiplex ligation-dependent probe amplification (MLPA) ratio plots validating copy-number measurement of PPP2R3B (4 probes) and the two telomeric genes GTPBP6 and PLCXD1 (one probe each), to the left of each figure and less than 150 bp in length; control probes of greater than 160 kb targeting genes of known normal copy number across the genome are shown to the right at greater than 150 bp size. A representative example of normal copy number for all genes (c), and of a duplication of PPP2R3B and GTPBP6 and PLCXD1 in a CMN patient (red dots) (d). While this method was able to validate the array CGH findings, it was not as robust as the targeted NGS panel for novel discovery of copy-number changes, likely due to the repetitive, GC-rich, and polymorphic nature of the region studied. Low-copy repeats at Xp22.33. (e) The upper panel depicts a regional similarity search across Xp22.33 with YASS software (http://bioinfo.cristal.univ-lille.fr/yass/index.php) both forward (green) and backward (red) revealing three segmental duplications (LCRA, LCRB, and LCRC) 5’ of PPP2R3B and a high density of SINE and LINE repeats. No segmental duplications are detected 3’ to PPP2R3B before SHOX. The assembly gaps (red), local genes (purple), and the homology region (orange) with the Y chromosome are indicated.