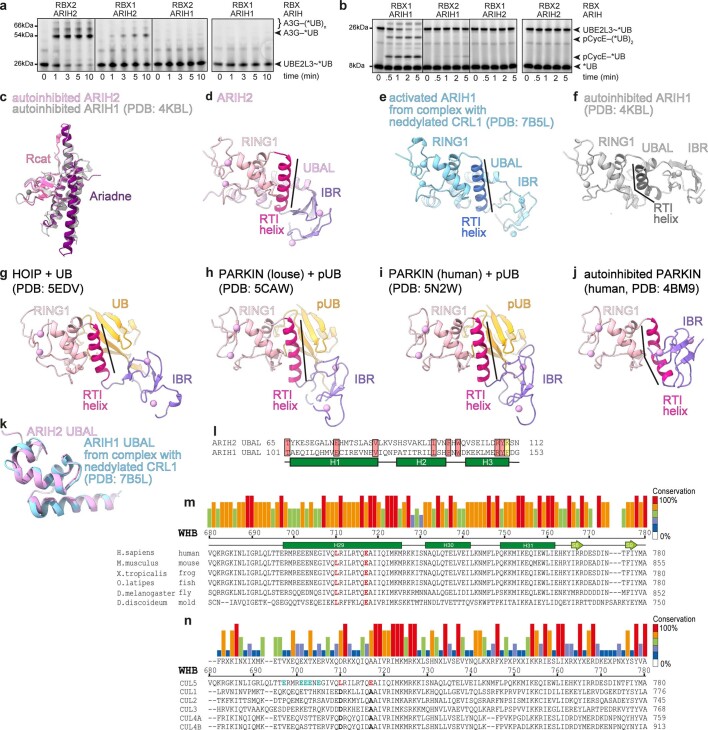
Extended Data Fig. 8. Neddylated CUL5-RBX2-ARIH2 and neddylated CUL1-RBX1-ARIH1 specificity.
a, Fluorescent scan of gels showing neddylated CRL5Vif-CBFβ and ARIH-family RBR E3-dependent *UB transfer from UBE2L3 through E3-E3 cascade for ubiquitylating A3G substrate, testing roles of ARIH E3 identity (ARIH1 or ARIH2) and RBX identity in neddylated CRL5Vif-CBFβ generated from Vif-CBFβ-ELOBC and either neddylated CUL5-RBX2 or CUL5-RBX1 (Source Data Extended Data Fig. 8). The data are representative from N = 2 independent experiments. b, Assays performed as in a, except with neddylated CRL1FBXW7∆D and phosphopeptide derived from Cyclin E (pCycE) as substrate (Source Data Extended Data Fig. 8). c, Superposition of Ariadne and Rcat domains in autoinhibited ARIH2 and ARIH1 (PDB ID: 4KBL25). d, UBAL–RING1–RTI-helix–IBR domains (that is, E2~UB-binding platform) of ARIH2 crystal structure. e-j, RING1–RTI-helix–IBR domains (that is, E2~UB-binding platform) of different RBRs (neddylated CRL1 bound ARIH1 PDB ID: 7B5L23, autoinhibited ARIH1 PDB ID: 4KBL25, HOIP PDB ID: 5EDV34, louse PARKIN PDB ID: 5CAW42, human PARKIN PDB ID: 5N2W43, autoinhibited human PARKIN PDB ID: 4BM926) aligned over their RING1 domains as in d. k, Superposition of the UBAL domain of ARIH2 crystal structure with that of a neddylated CRL1-bound ARIH1 (PDB ID: 7B5L23). l, Alignment of UBAL domain sequences of human ARIH2 and ARIH1. Secondary structures are indicated by rectangles for helices. Residues identical between the two are shaded in rose. ARIH1 F150 and ARIH2 K110 are shaded in yellow. m, Alignment of WHB domain sequences for CUL5 from the indicated organisms. Secondary structures based on crystal structure of unneddylated CUL512 are indicated by rectangles for helices, arrows for β-strands. Degree of conservation is indicated by color-coded bars above. L710 and E717, which configure noncovalent interactions with covalently linked NEDD8 are highlighted in red. n, Alignment of WHB domain sequences from human CUL5, CUL1, CUL2, CUL3, CUL4A, and CUL4B. Degree of conservation is indicated by color-coded bars above. In CUL5 sequence, L710 and E717, which configure noncovalent interactions with covalently linked NEDD8 are highlighted in red. The corresponding aspartate and alanine residues from CULs1–4 are highlighted black. CUL5 glutamates that are candidates for securing H29-helix in unneddylated CUL5 and that were mutated to lysines in E-to-K mutant are highlighted in cyan.