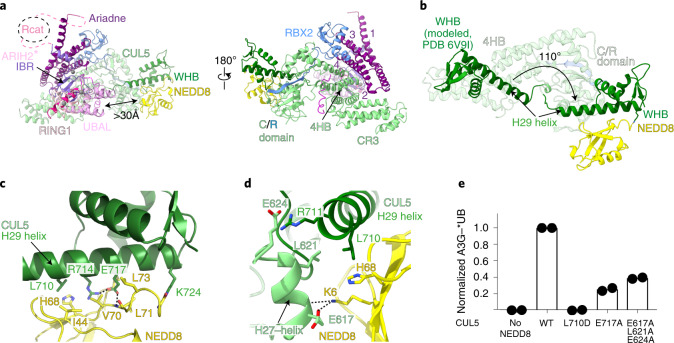
Fig. 2. NEDD8 conformational activation of CUL5-RBX2.
a, Structure of ARIH2* complex with neddylated CUL5 (spanning from CR3 domain to C terminus)-RBX2 is shown with domains colored as in Fig. 1. Black arrow indicates the >30 Å distance between the ARIH2* UBAL domain and NEDD8. Zinc atoms as spheres, and Ariadne domain helices numbered. b, Structural transition of CUL5 H29-helix and WHB domain between the unneddylated CUL5-RBX2 and neddylated CRL5Vif-CBFβ-A3C-ARIH2* complex (ARIH2* and RBX2 RING domain are not shown for simplification). To visualize the relative position of the unneddylated CUL5 H29-helix and WHB domain (dark green), the region encompassing the CR3, 4HB and C/R domains from unneddylated CUL5-RBX2 (ref. 22) was superimposed on the neddylated CUL5-RBX2-ARIH2* structure, but is not shown for simplification. In unneddylated CUL5, the H29-helix encompasses residues 697–725, whereas the first turn is unfolded and the H29-helix encompasses residues 700–725 in neddylated CUL5. c, Close-up of interactions surrounding the covalent isopeptide linkage between NEDD8 (yellow) and CUL5 H29-helix portion of the WHB domain (dark green). d, Three-way interface between NEDD8, its covalently linked CUL5 H29 helix and WHB domain, and a CUL5 surface from the C/R domain (relative to c, rotated 80° in x and 40° in y). e, ARIH2-catalyzed fluorescent UB (*UB) transfer to A3G in 10 min, mediated by WT unneddylated (no NEDD8), WT neddylated CRL5Vif-CBFβ (WT) or versions with indicated mutations of CUL5 residues making noncovalent interactions with NEDD8. N = 2 independent experiments. For samples from same experiment, gels were processed in parallel (Source Data Fig. 2).