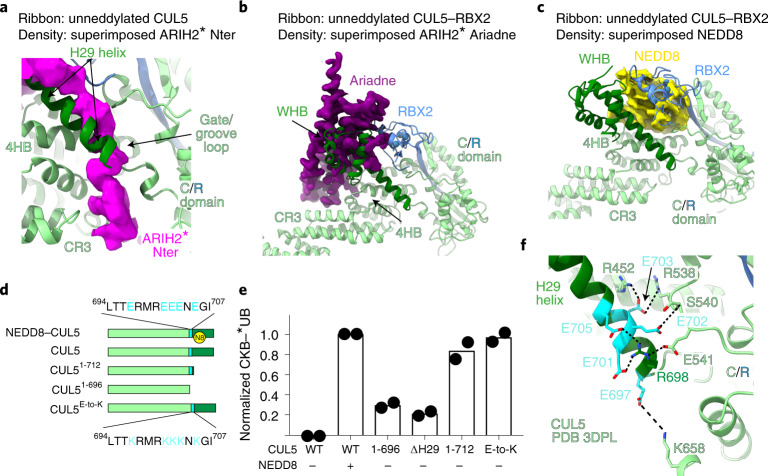
Fig. 4. ARIH2 binds surfaces exposed by NEDD8-mediated CUL5-RBX2 conformational changes.
a, ARIH2’s N-terminal region binds a groove that in unneddylated CUL5-RBX2 is occupied by CUL5’s gate/groove loop and H29-helix, shown by overlaying cryo-EM density for ARIH2* N-terminal region (magenta) from the complex with neddylated CRL5Vif-CBFβ-A3C on unneddylated CUL5 (green)-RBX2 (ref. 22) aligned over the C/R domain. b, ARIH2’s Ariadne domain binds a surface that in unneddylated CUL5-RBX2 is occupied by CUL5’s WHB and RBX2’s RING domains, shown by overlaying cryo-EM density for ARIH2* Ariadne domain (purple) from the complex with neddylated CRL5Vif-CBFβ-A3C on unneddylated CUL5-RBX2 (green and blue)22 aligned over the C/R domain. c, CUL5-linked NEDD8 binds a surface that in unneddylated CUL5-RBX2 is occupied by RBX2’s RING domain, shown by overlaying cryo-EM density for NEDD8 (yellow) linked to CUL5 in complex with ARIH2* on WHB domain of unneddylated CUL5-RBX2 (green and blue)22. d, Schematics of CUL5 constructs used to test effects of removing regions of unneddylated CUL5 shown in a and b as blocking ARIH2-binding sites. CUL5’s H29-helix and WHB domain are indicated in dark green. Glutamates securing position of the unneddylated CUL5 H29-helix are indicated in cyan in WT CUL5. Lysine substitutions in CUL5 ‘E-to-K’ mutant are shown below. e, ARIH2-catalyzed fluorescent UB (*UB) transfer to CKB in 10 min, mediated by unneddylated or neddylated WT CRL5ASB9, or with indicated mutant versions of unneddylated CUL5. N = 2 independent experiments. For samples derived from the same experiment, gels were processed in parallel (Source Data Fig. 4). f, Structure of unneddylated CUL5 (ref. 12) showing interaction network of glutamates in H29-helix.