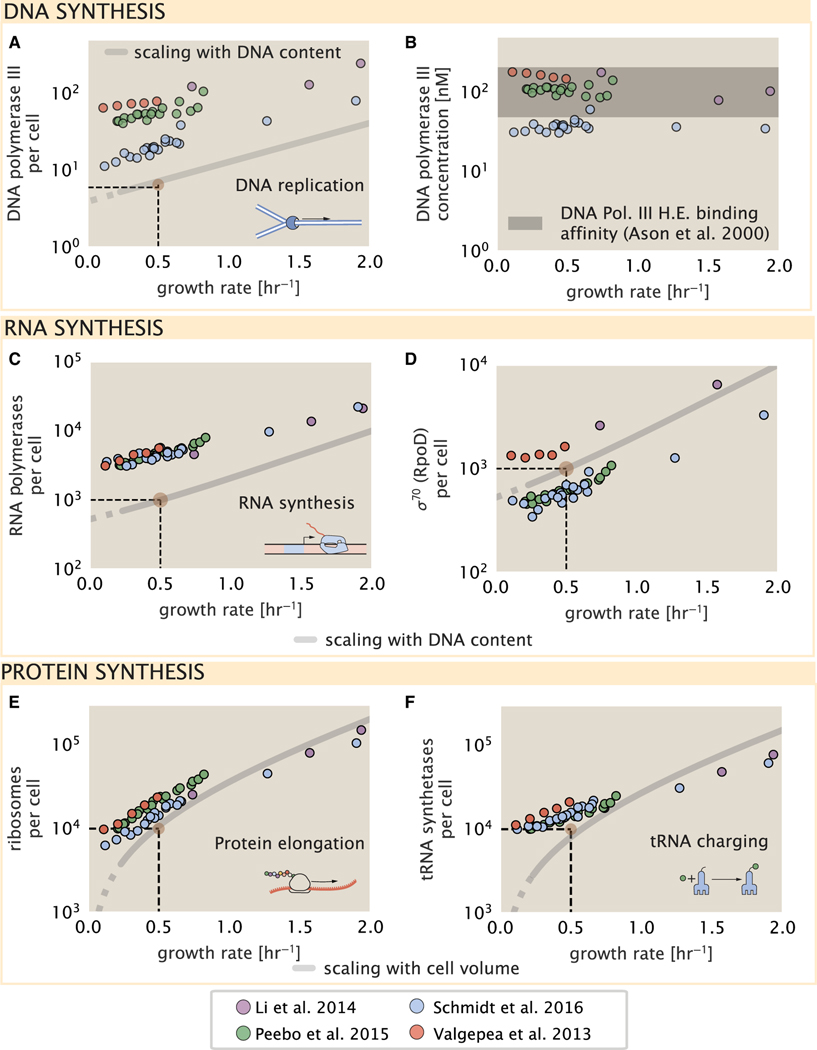
Figure 5. Processes of the central dogma.
(A) The minimum number of DNA polymerase holoenzyme complexes needed to facilitate replication of the genome. Points correspond to the total number of DNA polymerase III holoenzyme complexes ([DnaE]3[DnaQ]3[HolE]3[DnaX]5[HolB] [HolA][DnaN]4[HolC]4[HolD]4) per cell.
(B) The effective concentration of DNA polymerase III holoenzyme (see supplemental information section “estimation of cell size and surface area” for calculation of cell size). Shaded region corresponds to the range of KD values measured by Ason et al. (2000), from 50 to 200 nM.
(C) The number of RNA polymerase core enzymes, with measurements corresponding to the average number given a subunit stoichiometry of [RpoA]2[R-poC][RpoB].
(D) The abundance of σ70 as a function of growth rate along with the same prediction from (C).
(E) Number of ribosomes required to synthesize 109 peptide bonds with an elongation rate of 15 peptide bonds per second.
(F) Number of tRNA synthetases that will supply the required amino acid demand. The sum of all tRNA synthetases copy numbers are plotted ([ArgS], [CysS], [GlnS], [GltX], [IleS], [LeuS], [ValS], [AlaS]2, [AsnS]2, [AspS]2, [TyrS]2, [TrpS]2, [ThrS]2, [SerS]2, [ProS]2, [PheS]2[PheT]2, [MetG]2, [lysS]2, [HisS]2, [GlyS]2[GlyQ]2). Dashed black lines indicate order of magnitude estimate needed at a growth rate of ≈0.5 per h (light-brown point), while the gray line accounts for the growth rate dependence changes in cell size and doubling time. Dashed region of gray line represents growth rates with a doubling time ≥ 3 h where protein maintenance costs may be important but are not considered.