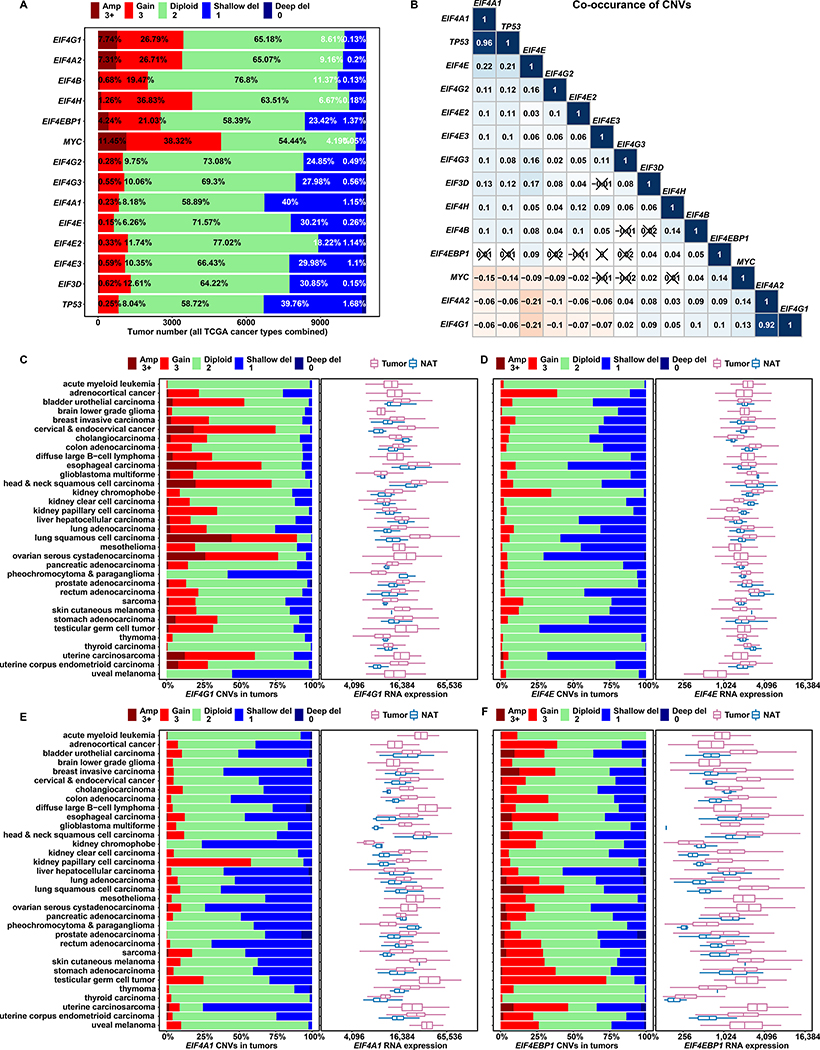
Figure 1. Among Initiation Factor Genes, EIF4G1 Is Frequently Amplified and Overexpressed in Tumors.
(A) The stacked bar plot shows overall CNV statuses for translation initiation genes in all tumors combined from 33 TCGA cancer types. We used TCGA groupings of CNV value estimates that were derived from whole genome microarray data by the GISTIC2 method. The estimated gene-level CNV values were grouped with thresholds 3+, 3, 2, 1, 0, to represent high-level copy number gain (amplification), low-level copy number gain (duplication), diploid, shallow (possibly heterozygous) deletion, or deep (possibly homozygous) deletion, respectively. Percentage contributions of each group are labeled on the bars.
(B) The matrix plot shows Pearson correlation coefficients for translation initiation gene CNVs. For each gene, a list of estimated CNV values for 10,845 tumors (all TCGA study groups) is correlated with the corresponding list for another gene. Each cell is colored based on the magnitude of the resulting coefficient, and cells with ‘X’ indicate statistical insignificance (p > 0.05, p values not shown). Aside from identity relationships (topmost diagonal cells), strong co-occurrence (dark blue) is evident for TP53/EIF4A1 and EIF4G1/EIF4A2 – gene pairs with neighboring chromosomal locations.
(C to F) For each cancer study group, a stacked bar plot shows CNV status for a translation initiation gene (marked at the bottom of each plot), and a box and whisker plot shows corresponding mRNA expression of the same gene in tumor samples and Normal Adjacent Tissues (NATs). The X axes of box plots represent normalized gene level expression (transcripts per million) in log2 scale. TCGA uses the same bioinformatics pipeline to process and normalize RNA-Seq data from different cancer study groups, to minimize batch effects of sequencing data processing