Fig. 2. Evaluation of hyPE2 linker variants.
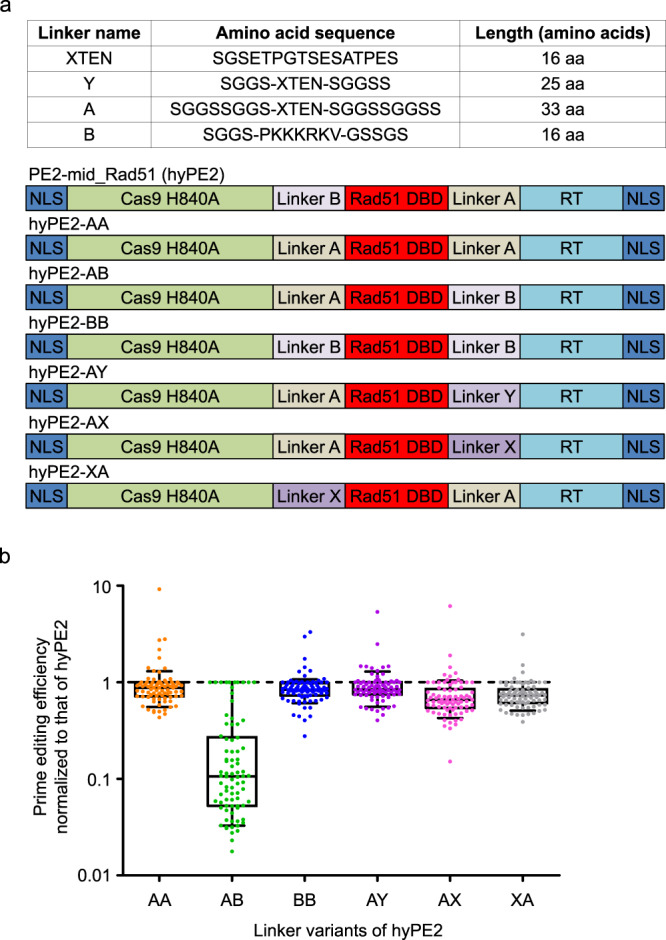
a Detailed information about the linkers that were used and the structures of the hyPE2 linker variants. DBD DNA-binding domain, NLS nuclear-localization signal, RT reverse transcriptase. b Prime-editing efficiencies of hyPE2 variants normalized to the efficiency of hyPE2 at the same target sequences, which had been lentivirally integrated in HEK293T cells. Adjusted fold increases are shown on the y axis. Data of minimum-to-maximum values are presented. For the boxes, the top, middle, and bottom lines represent the 25th, 50th, and 75th percentiles, respectively. The whiskers indicate the 10th- and 90th-percentile values. The number of pegRNAs n = 82. Source data are provided as a Source Data file.
