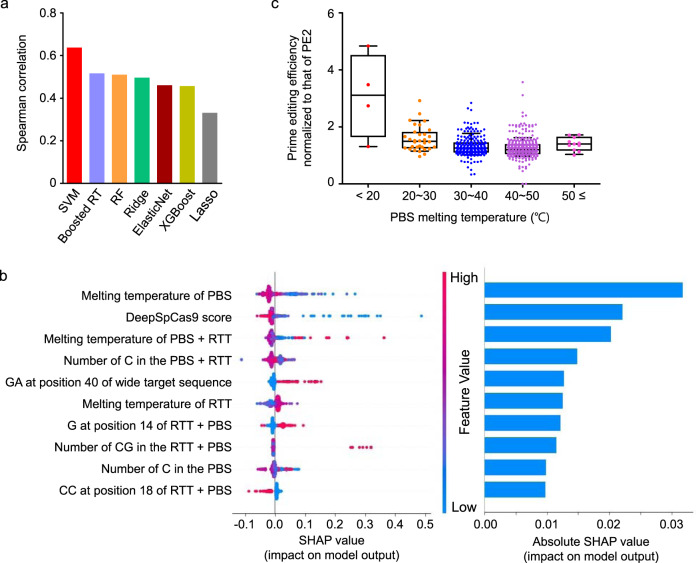
Fig. 4. Development and evaluation of PEselector, a computational model that predicts the fold increase of hyPE2-induced prime-editing efficiencies as compared to those of PE2.
a Comparison of Spearman correlation coefficients between prediction models. SVM, support-vector machine; Boosted RT, gradient-boosted regression tree; RF, random forest; Ridge, L2-regularized linear regression; ElasticNet, L1L2-regularized linear regression; XGBoost, extreme-gradient boosting; Lasso, L1-regularized linear regression. b The top ten features associated with hyPE2 activity as compared with PE2 activity determined by Tree SHAP (XGBoost classifier). The dot colors indicate the high (red) or low (blue) values of the relevant feature for each pegRNA. Overlapping points are slightly separated in the y-axis direction, so that density is apparent. c Dependence of the fold increase in hyPE2 prime-editing efficiency relative to that of PE2 on the primer-binding site (PBS) melting temperature. Editing efficiencies for hyPE2 and PE2 were determined at the same target sequences, which had been lentivirally integrated in HEK293T cells (library B). Data points are overlaid. The number of pegRNAs n = 4 (<20 °C), 32 (20–30 °C), 236 (30–40 °C), 348 (40–50 °C), and 11 (≥50 °C). Data of minimum-to-maximum values are presented. For the boxes, the top, middle, and bottom lines represent the 25th, 50th, and 75th percentiles, respectively. The whiskers indicate the 10th- and 90th-percentile values. Source data are provided as a Source Data file.