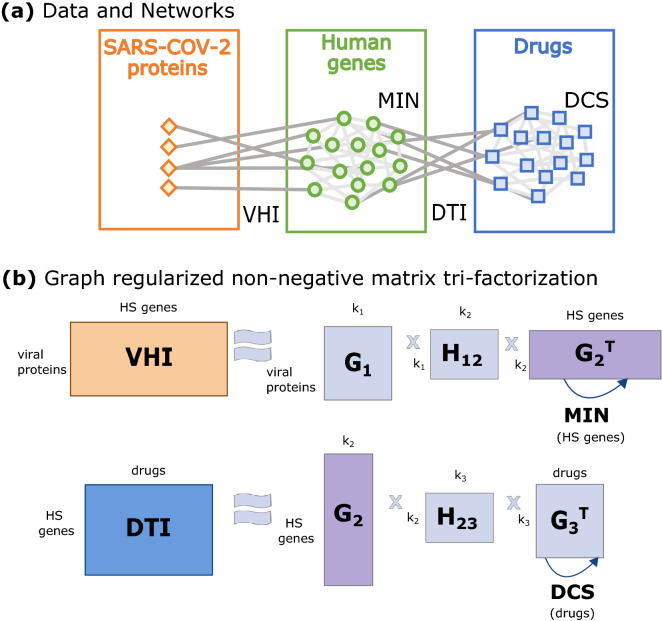
Figure 1.
Illustration of the data and framework. (a) Schematic illustration of datasets used in this study. Three data types are represented: SARS-CoV-2 proteins (in orange), human genes (in green) and drugs (in blue). Two relational datasets connect different types of data: virus-host protein–protein interactions (VHIs) and drug-target interactions (DTIs). Network structural knowledge from these data types is contained in the molecular interaction network (MIN) and the drug chemical similarity (DCS) network. (b) Graph-regularized non-negative matrix tri-factorization (GNMTF) used for fusing the VHIs, DTIs, MIN and DCS networks. The matrix factor is shared across decompositions to simultaneously decompose the VHI and DTI networks. Network structure (topology) information from the MIN and DCS networks are incorporated into the data fusion by using two regularization terms (illustrated by arcs with arrows). The parameters , and indicate the numbers of clusters of viral proteins, human genes and drugs, respectively.