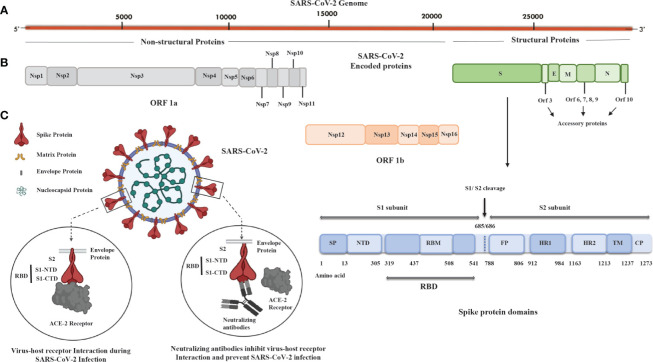
Figure 1.
SARS-CoV-2 genome, encoded proteins, and basic mechanism of virus fusion and entry. (A) Illustration of the SARS-CoV-2 genome that is around 30 kb in size and has a 5’ cap and 3’ poly A tail. (B) SARS-CoV-2 proteins: nonstructural proteins (Nsp), ORF1a, and ORF1b (Nsp1-Nsp16) and structural proteins such as spike (S), envelope (E), membrane (M), and nucleocapsid (N) with spike protein having 1273 aa (~180 kDa) containing signal peptide (SP), N-terminal domain (NTD), receptor binding motif (RBM) in receptor binding domain (RBD), fusion peptide (FP), heptad repeat (HR)-1, HR-2, transmembrane domain (TM), and cytoplasmic tail (CP) domains. There are accessory proteins Orf3; Orf6,7,8,9; and Orf10 located in between the S, E, M, and N proteins. (C) The spike protein subunit 1 (S1; aa 14-685) of SARS-CoV-2 consists of the RBD domain divided into two parts: S1 N-terminal domain (S1-NTD) and S1 C-terminal domain (S1-CTD) and spike protein subunit 2 (S2; aa 686-1273) containing TM and CP domains. S1-CTD interacts with the angiotensin-converting enzyme-2 (ACE-2) receptor of host cells and facilitates fusion and entry of virus. The neutralizing antibody against S1-CTD blocks the entry of the SARS-CoV-2 into host cells. ORF1a; Open reading frame 1a, ORF1b; Open reading frame 1b, aa; Amino acid.