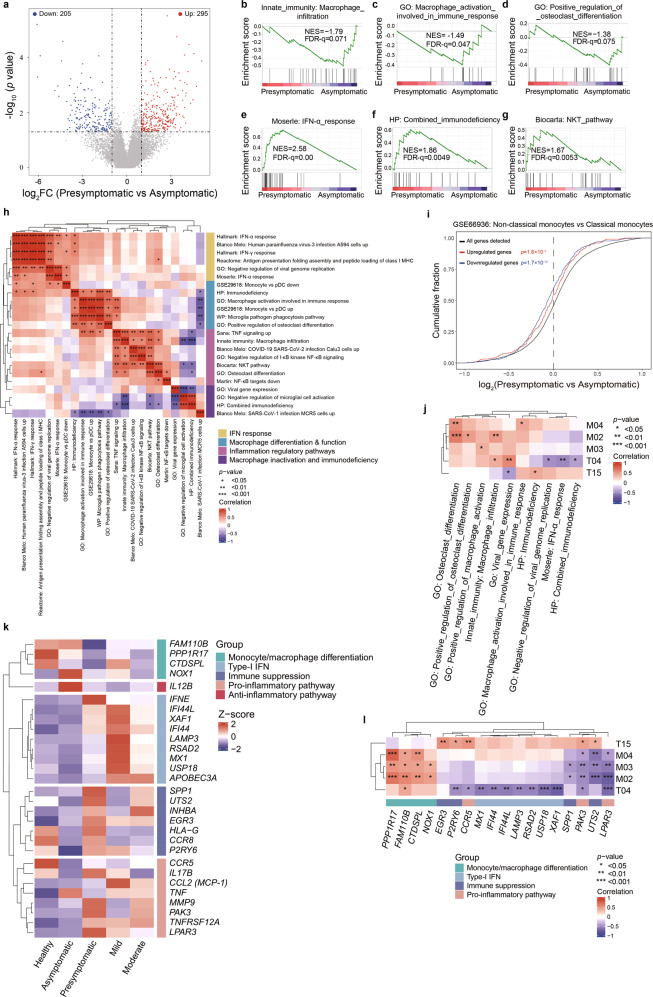
Fig. 4. A few crucial PBMC-related signaling pathways differentially function in the asymptomatic and presymptomatic cases.
a Volcano plot showing the genes differentially expressed (fold change ≥ 2, P < 0.05) in PBMCs between the presymptomatic versus asymptomatic cases. b–g GSEA of the expression profile of the presymptomatic or asymptomatic cases using macrophage infiltration-associated signature (b), macrophage activation-involved immune response-associated signature (c), osteoclast differentiation-associated signature (d), IFN-α response-associated signature (e), combined immunodeficiency-associated signature (f) and NKT pathway-associated signature (g). h Unsupervised hierarchical clustering of the Spearman correlation matrix of the gene sets differentially expressed between the asymptomatic and presymptomatic cases among SSIS groups. Gene set signature scores were calculated from mRNA expression as predefined linear combinations (weighted averages) of biologically relevant genes. i Empirical cumulative distribution function for the change in expression (log2 values) of all genes (black line) expressed in PBMCs of the presymptomatic subjects (change relative to that in PBMCs of asymptomatic subjects) and for subsets of genes upregulated (red line) or downregulated (blue line) in non-classical monocytes as compared with classical monocytes (GSE66936). j Heatmap showing Spearman correlations between the frequencies of indicated clusters and the enrichment of selected gene sets in the four SARS-CoV-2pos groups. k Heatmap showing a number of genes differentially expressed between the presymptomatic and asymptomatic subjects. The functional categories of the genes are indicated. l Heatmap showing Spearman correlations between the frequencies of indicated clusters and the mRNA levels of selected genes in the four SARS-CoV-2pos groups. The functional categories of the individual genes are indicated.