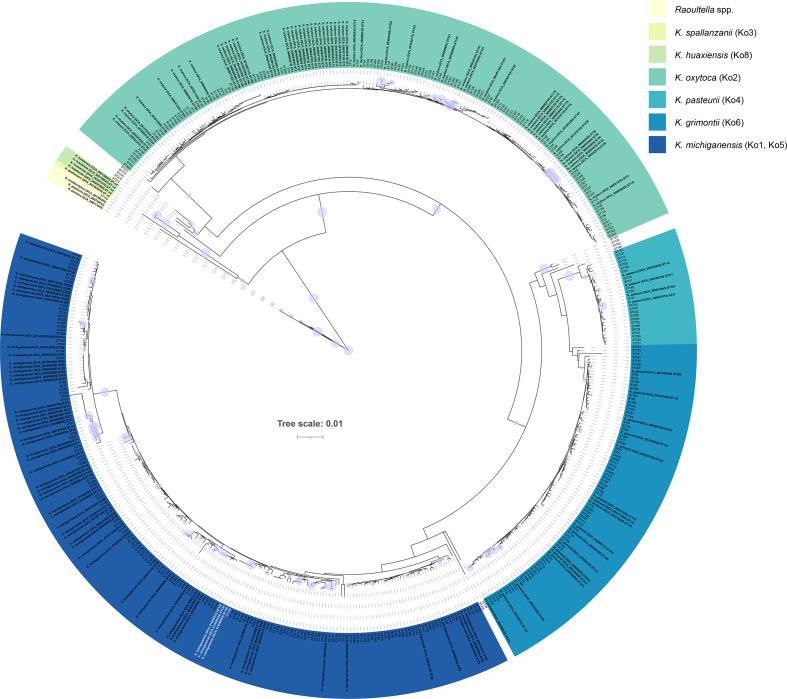
Fig. 1.
Sequence types within the K. oxytoca MLST scheme can be used to distinguish members of the K. oxytoca complex. The three K. michiganensis clinical isolates (all ST138) characterized in this study are shown in white. The phylogenetic tree (neighbour joining, Jukes Cantor) was generated using concatenated nucleotide sequences of housekeeping genes (gapA–infB–mdh–pgi–phoE–rpoB–tonB) used in the K. oxytoca MLST scheme [8]. The purple circles represent bootstrap values ≥80 % (based on 1000 replications); the larger the circle, the higher the bootstrap value. Scale bar, average number of nucleotide substitutions per position. The full list of MLST sequence types and their species affiliations are available in Table S2.