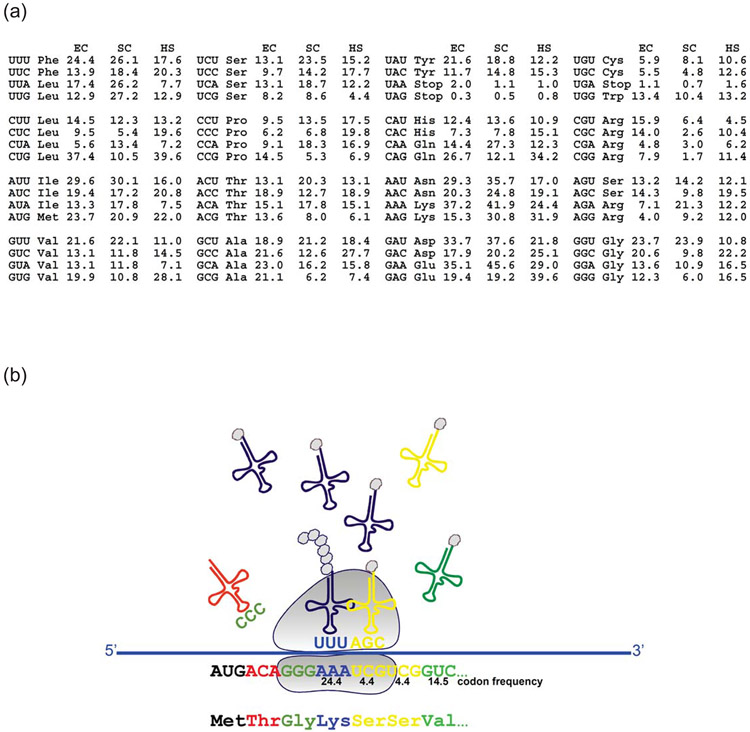
Figure 1. Genetic code redundancy and non-uniform/non-random codon utilization shape codon usage bias and govern non-uniform translation.
(a) Codon usage bias in Escherichia coli (EC), Saccharomyces cerevisiae (SC) and Homo sapiens (HS) (https://www.kazusa.or.jp/codon/). These examples reveal substantial differences in usage of codons between the three species. Frequency per thousand codons is shown. (b) Preferentially used (frequent) codons are translated faster than infrequently used (rare) codons due to the more ready availability (during translation) of the corresponding frequent cognate tRNAs. Rare codons, as a rule, lead to a substantial increase in ribosome residence time due to an increased waiting period of the ribosome for a cognate tRNA. A simplified scheme is shown (omitting the elongation factor 1A, which promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis). Colors of tRNAs/codons correspond to the differential frequency of their usage.