Abstract
Objectives
To compare the demographics, clinical characteristics and severity of patients infected with nine different SARS-CoV-2 variants, during three phases of the COVID-19 epidemic in Marseille.
Methods
A single centre retrospective cohort study was conducted in 1760 patients infected with SARS-CoV-2 of Nextstrain clades 20A, 20B, and 20C (first phase, February–May 2020), Pangolin lineages B.1.177 (we named Marseille-2) and B.1.160 (Marseille-4) variants (second phase, June–December 2020), and B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma) and A.27 (Marseille-501) variants (third phase, January 2021-today). Outcomes were the occurrence of clinical failures, including hospitalisation, transfer to the intensive-care unit, and death.
Results
During each phase, no major differences were observed with regards to age and gender distribution, the prevalence of chronic diseases, and clinical symptoms between variants circulating in a given phase. The B.1.177 and B.1.160 variants were associated with more severe outcomes. Infections occurring during the second phase were associated with a higher rate of death as compared to infections during the first and third phases. Patients in the second phase were more likely to be hospitalised than those in the third phase. Patients infected during the third phase were more frequently obese than others.
Conclusion:
A large cohort study is recommended to evaluate the transmissibility and to better characterise the clinical severity of emerging variants.
Keywords: COVID-19, SARS-CoV-2, Variant, Mutation, 501Y, Marseille
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 in Wuhan in the Hubei province of China and causes the disease known as coronavirus disease 2019 (COVID-19). Since its emergence, SARS-CoV-2 has spread worldwide, and COVID-19 has caused the unprecedented disruption of human society. By 15 August 2021, the pandemic had infected over 205 million people and has led to the deaths of more than four million patients (Worldometers, 2021). The presentation of the disease ranges from asymptomatic to severe and fatal forms, especially among older and vulnerable populations (WHO, 2021; Gautret et al., 2020).
The diversity of the SARS-CoV-2 was initially reported to be very low (Fauver et al., 2020). However, variants, which differ from all other strains by sets of several (at least 5) mutations, in contrast with mutants that result from the progressive accumulation of mutations during viral replication and spread, have been reported since summer 2020, including in our geographical area (Fournier et al., 2021a). In late December 2020, new variants of concern emerged, particularly those first described in the UK (named B.1.1.7 in Pangolin classification or alpha in WHO classification), South Africa (B.1.351 or beta), Brazil (P.1 or gamma) and India (B.1.167.2 or delta; Faria et al., 2021; European Centre for Disease Prevention and Control, n.d.), which have become major concerns. Indeed, their spike protein, which is the major target of immune responses elicited by previous infections or vaccination, may harbour several amino acid substitutions, including N501Y and E484K, or deletions that confer decreased sensitivity to antibodies (Wang et al., 2021).
Preliminary reports in the UK suggested that the alpha variant is more transmissible than previously circulating viruses, with increase in transmissibility estimated to be up to 75% (European Centre for Disease Prevention and Control, 2021). The beta variant has also spread rapidly in South Africa. In addition, both variants have spread to several countries located in six WHO regions (https://nextstrain.org/ncov/global; PAHO, 2021). The SARS-CoV-2 gamma variant, initially identified in Manaus, Amazonas State, Brazil has also spread to various countries (PAHO, 2021). The delta variant was first detected in India in late 2020. It is thought to be partly responsible for India's second wave of the pandemic beginning in February 2021. It later contributed to a third wave in Fiji, the United Kingdom and South Africa (Callaway, 2021). To date, this variant affected 105 countries and more than 250,000 sequences were recorded (https://cov-lineages.org/global_report_B.1.617.2.html).
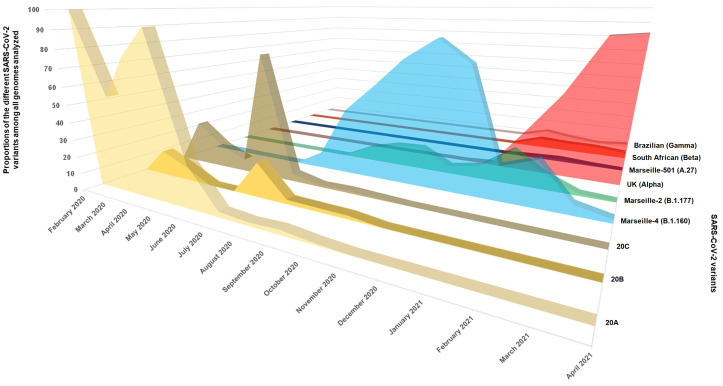
In Marseille, the first case of COVID-19 was recorded at the Institut Hospitalo-Universitaire Méditerranée Infection (IHU) on 27 February 2020 and the epidemic was characterised by three major phases. The first (phase 1) started in February and almost ended in May (Fournier et al., 2021a; Colson et al., 2021a). The second phase occurred suddenly at the end of June and lasted until December 2020. The third phase started in January 2021 and ended in June 2021. SARS-CoV-2 genomes were sequenced over time to characterise the genetic diversity of SARS-CoV-2. During the first phase of the epidemic, viruses of lineages 20A, 20B and 20C that are closely related to the initial Wuhan-Hu-1 isolate were predominantly circulating (Colson et al., 2021a; Colson et al., 2021b). In January 2021, 14 variants, with clearly distinct genomic patterns, concomitantly or successively spread in the Marseille area (Colson et al., 2021a; Dao et al., 2021a), with three variants, including Marseille-1 (B.1.5.12.1 Pangolin lineage), Marseille-2 (B.1.177) and Marseille-4 (B.1.160), successively predominating (Colson et al., 2021a; Fournier et al., 2021b). The first case of a variant harbouring N501Y substitution (N501YV) was diagnosed in our institute on 31 December 2020, starting the third phase of the epidemic in Marseille. Up to June 2021, the alpha variant was the main variant circulating during this third phase, followed by the gamma variant, the Marseille-501 variant (A.27 Pangolin lineage; Colson et al., 2021c) and the beta variant (Fig. 1 ). In a preliminary study, we observed that patients infected with 20A viruses of the first phase, B.1.5.12.1 and B.1.160 variants, and variants harbouring N501Y mutation presented different patterns of symptoms and severity (Fournier et al., 2021a; Colson et al., 2021a; Dao et al., 2021a). In this paper, we compare the demographics, clinical characteristics and severity of patients infected with nine different SARS-CoV-2 variants, during the three phases of the COVID-19 epidemic in Marseille.
Fig. 1.
Weekly distribution of SARS-CoV-2 genotypes.
2. Material and methods
2.1. Data source
We conducted a single centre retrospective cohort study at the IHU Méditerranée Infection, Marseille (France), which is part of the network of public hospitals in Marseille (AP-HM). All available SARS-CoV-2 genome sequences obtained by our laboratory between March 2020 and April 2021 were reviewed. Patients infected with 20A, 20B, 20C viruses (circulating during the first phase of epidemic), B.1.177 or B.1.160 variants (circulating during the second phase), or different variants (N501YV) harbouring the N501Y substitution within the spike protein (circulating during the third phase of pandemic), were selected (Fig. 1). Additional patients diagnosed with SARS-CoV-2 by real-time reverse-transcription PCR (qPCR) were included for whom viral genotype was determined by partial spike gene sequencing as previous described (Colson et al., 2021c) or the Applied Biosystems TaqPath COVID-19 kit (Thermo Fisher Scientific, Waltham, USA) or in house variant-specific qPCR as previously described (Fournier et al., 2021a; Bedotto et al., 2021). A second filter was applied to include only patients with information available on clinical status and follow-up. The B.1.5.12.1 variant that reached a very weak peak but represented up to 100% of infections during part of the month of July and then disappeared after a month-and-a-half has been described elsewhere (Colson et al., 2021a).
2.2. Patients
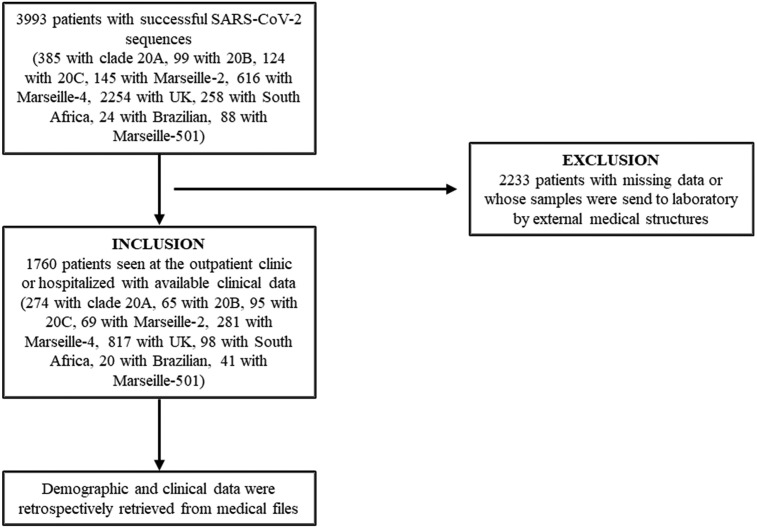
The IHU received patients or asymptomatic contacts directly presenting for SARS-CoV-2 testing or samples sent from other wards in the Marseille Public University hospitals (AP-HM; particularly in temporarily dedicated COVID-19 units and in intensive care units), or from laboratories outside the AP-HM. Most of the positive patients sampled at IHU were followed-up in the day clinic or were hospitalised in the Infectious Diseases department of the IHU, according to the severity of the disease. No detailed information was available for patients whose samples were sent to the IHU laboratory (Fig. 2 ).
Fig. 2.
Flow-chart of study.
In this study, we only included 1760 patients who were seen at our institute in the day clinic or who were hospitalised in the conventional infectious disease units or other wards within the AP-HM. Demographic and clinical data including comorbidities were retrospectively retrieved from medical files including, notably, the main symptoms, in/outpatient status, transfer to intensive care unit (ICU), and death. At the time of writing, all patients had recovered and been discharged from hospital or had died. Because death could possibly occur long-time post-discharge, mortality was investigated through the national data on COVID-19 related mortality (at least 30 days post-discharge; Institut National de la Statistique et des Etudes Economiques (INSEE), 2021). Patients with missing information were mostly patients whose samples were send to our laboratory by external medical facilities and who were therefore excluded.
2.3. Genome sequencing and assembling
All samples that were positive for SARS-CoV-2, identified by real-time PCR (Amrane et al., 2020) with a cycle threshold (Ct) value < 30, were processed for next-generation sequencing over time to characterise the genetic diversity of the virus. Whole genome sequencing was performed as previously described (Colson et al., 2021a) from viral RNA extract obtained from 200 μL of nasopharyngeal swab fluid by reverse transcription by SuperScript IV (ThermoFisher Scientific, Waltham, MA, USA), cDNA second strand synthesis using Klenow Fragment DNA polymerase (New England Biolabs, Beverly, MA, USA), and generated DNA purification with Agencourt AMPure XP beads (Beckman Coulter, Villepinte, France). Genome next-generation sequencing was performed using various techniques throughout the period of the SARS-CoV-2 pandemic: with the Illumina technology using the Nextera XT paired end strategy on MiSeq instruments between February 2020 and mid-March 2021 (Illumina Inc., San Diego, CA, USA), as previously described (Fournier et al., 2021a); with the Oxford Nanopore technology on a GridION instrument between mid-March and mid-April 2021 (Oxford Nanopore Technologies Ltd., Oxford, UK), as previously described (Colson et al., 2020), after viral RNA reverse-transcription using SuperScript IV (ThermoFisher Scientific) then cDNA second strand synthesis with LunaScript RT SuperMix kit (New England Biolabs) and synthesized cDNA amplification using a multiplex PCR protocol with ARTIC nCoV-2019 V3 Panel primers (Integrated DNA technologies, Coralville, IA, USA) according to the ARTIC procedure (https://artic.network/); and with the Illumina COVIDSeq protocol on a NovaSeq 6000 instrument since mid-April 2021 (Illumina Inc., San Diego, CA, USA) following the manufacturer's instructions.
Genome assembly was performed using the CLC Genomics workbench v.7 software by mapping on the SARS-CoV-2 genome GenBank Accession no. NC_045512.2 (Wuhan-Hu-1 isolate). Recovered genomes were compared to sequences from the GISAID database (https://www.gisaid.org/). The phylogenetic analysis was performed using the nextstrain/ncov tool (https://github.com/nextstrain/ncov) that uses the IQ-TREE software (Minh et al., 2020) for phylogenetic tree building then the Auspice software for tree visualisation (https://docs.nextstrain.org/projects/auspice/en/latest/releases/v2.html).
2.4. Statistics
Statistical analyses were carried out using R (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2020. URL: https://www.Rproject.org/) and Stata version 15.1 (http://www.stata.com). Qualitative variables were presented by percentage. We used two approaches to conduct the analysis. The first one examined the demographics and clinical profiles of patients infected with different viral mutants or variants during each phase of the epidemic: 20A vs. 20B vs. 20C (first phase), B.1.177 vs. B.1.160 (second phase) and alpha vs. beta vs. gamma vs. A.27 (third phase). The second approach evaluated the difference in clinical outcomes, including hospitalisation, transfer to the ICU, and death among patients infected with SARS-CoV-2 from various lineages during the three phases (first phase vs. second phase vs. third phase). Unadjusted associations between multiple factors and groups of variants or clinical outcomes were examined by univariate analysis. Variables with P values < 0.2 in the univariate analysis were included in the multivariate analysis (Bursac et al., 2008). The φ coefficient was used to test for multicollinearity among the independent variables. For pairs of variables that were highly correlated (absolute value of correlation coefficient > 0.7), only one variable was entered into the multivariate model. Multivariate analysis was performed using exact logistic regression. The results were presented by percentages and odds ratio (OR), with a 95% confidence interval (95% CI). A P-value < 0.05 was considered as statistically significant.
2.5. Ethics statement
Whole genome sequencing was performed on nasopharyngeal samples that were collected in the context of routine diagnosis. No additional samples were collected for this study. Clinical data were retrospectively retrieved from medical files and anonymised before analysis. Ethical approval was obtained from the Marseille Institutional Review Board and Ethics Committee (No. 2020–016-03).
3. Results
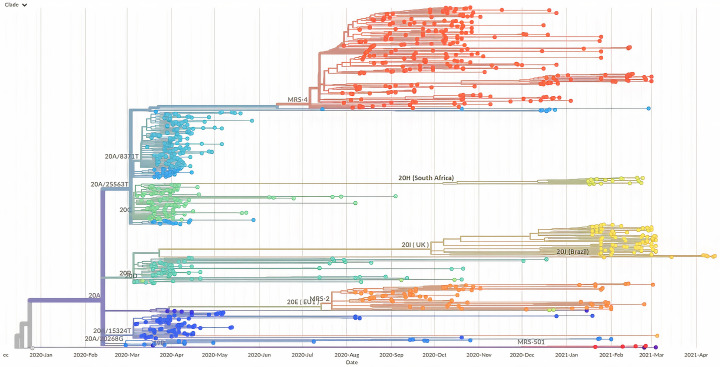
Between 29 February 2020 and 14 April 2021, we identified 3993 COVID-19 patients infected with nine variants, viral sequences being available from 3965 of them, including a full-length genomes in 1041 cases based on which a phylogeny was reconstructed (Fig. 3). Clinical data were available from 1760 of these 3993 patients who were treated at our institute, including patients infected with 20A (N = 274), 20B (N = 65), and 20C (N = 95) viruses during the first phase, with B.1.177 (N = 69) and B.1.160 (N = 281) variants during the second phase, and alpha (N = 817), beta (N = 98), gamma (N = 20) and A.27 (N = 41) variants during the third phase (Table S1).
Fig. 3.
Phylogeny reconstruction based on the SARS-CoV-2 genomes recovered from 1041 patients.
The genome of the original Wuhan-Hu-1 coronavirus isolate (GenBank accession no. NC_045512.2) was incorporated in the tree. Major SARS-CoV-2 variants are labelled. MRS-4, B.1.160 (Marseille-4); 20H (South Africa), B.1.351 (Beta); 20I (UK), B.1.1.7 (Alpha); 20 J (Brazil), P.1 (Gamma); MRS-2, B.1.1.177 (Marseille-2); MRS-501, A.27 (Marseille-501).
Patients with clinical data were older than patients without (P < 0.0001). No significant difference in gender was observed between the two groups (P = 0.65; Table S1).
3.1. Comparison of patients infected with 20A, 20B and 20C viruses during the first phase of the epidemic
Table 1 shows the characteristics of 434 patients infected with the three major clades circulating during the first phase of the COVID-19 epidemic in Marseille.
Table 1.
Characteristics of COVID-19 patients infected with clade 20A, 20B and 20C viruses during the first phase of the epidemic in Marseille (univariate and multivariate analysis).
| 20A |
20B |
20C |
20A vs. 20B |
20A vs. 20C |
20B vs. 20C |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n |
% |
n |
% |
n |
% |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
|
| N = 274 | N = 65 | N = 95 | Ref = 20B | Ref = 20C | Ref = 20C | |||||||
| Sociodemographic characteristic | ||||||||||||
| Mean age ± SD | 49.1 ± 20.4 | 39.2 ± 17.2 | 48.9 ± 16.7 | NA | NA | NA | NA | NA | NA | |||
| Age < 45 | 126 | 46.0 | 44 | 67.7 | 34 | 35.8 | ref | ref | ref | ref | ref | ref |
| 45–64 | 92 | 33.6 | 14 | 21.5 | 45 | 47.4 | 2.29 [1.18–4.43]; 0.01 | 2.44 [1.25–4.75]; 0.009 | 0.55 [0.33–0.93]; 0.03 | 0.59 [0.36–0.96]; 0.033 | 0.24 [0.11–0.51]; <0.0001 | 0.22 [0.10–0.48]; <0.0001 |
| ≥65 | 56 | 20.4 | 7 | 10.8 | 16 | 16.8 | 2.79 [1.19–6.58]; 0.02 | 3.38 [1.40–8.14]; 0.007 | 0.94 [0.48–1.85]; 0.87 | – | 0.34 [0.13–0.91]; 0.03 | 0.33 [0.12–0.91]; 0.033 |
| Gender | ||||||||||||
| Female | 152 | 55.5 | 33 | 50.8 | 49 | 51.6 | 0.83 [0.46–1.48]; 0.49 | NA | 0.85 [0.52–1.40]; 0.51 | NA | 1.03 [0.52–2.04]; 0.92 | NA |
| Male | 122 | 44.5 | 32 | 49.2 | 46 | 48.4 | ||||||
| Hypertension | 56 | 20.4 | 5 | 7.7 | 18 | 19.0 | 3.08 [1.17–10.27]; 0.02 | – | 1.10 [0.59–2.11]; 0.75 | NA | 0.36 [0.10–1.08]; 0.04 | – |
| Diabetes | 24 | 8.8 | 2 | 3.1 | 10 | 10.5 | 3.02 [0.72–27.01]; 0.12 | – | 0.82 [0.36–1.99]; 0.61 | NA | 0.27 [0.03–1.34]; 0.08 | – |
| Cancer | 13 | 4.7 | 1 | 1.5 | 2 | 2.11 | 3.19 [0.46–137.53]; 0.24 | NA | 2.32 [0.51–21.48]; 0.26 | NA | 0.73 [0.01–14.26]; 0.80 | NA |
| Chronic respiratory disease | 24 | 8.8 | 9 | 13.9 | 11 | 11.6 | 0.60 [0.25–1.55]; 0.21 | NA | 0.73 [0.33–1.73]; 0.42 | NA | 1.23 [0.42–3.49]; 0.67 | NA |
| Chronic heart disease | 33 | 12.0 | 6 | 9.2 | 8 | 8.4 | 1.35 [0.52–4.11]; 0.52 | NA | 1.49 [0.64–3.87]; 0.33 | NA | 1.11 [0.30–3.85]; 0.86 | NA |
| Obesity | 28 | 10.2 | 8 | 12.3 | 10 | 10.5 | 0.81 [0.33–2.17]; 0.62 | NA | 0.97 [0.43–2.33]; 0.93 | NA | 1.19 [0.38–3.59]; 0.73 | NA |
| Clinical signs | ||||||||||||
| Fever | 75 | 27.4 | 20 | 30.8 | 23 | 24.2 | 0.85 [0.46–1.62]; 0.58 | NA | 1.18 [0.67–2.13]; 0.55 | NA | 1.39 [0.64–3.00]; 0.36 | NA |
| Cough | 133 | 48.5 | 36 | 55.4 | 57 | 60.0 | 0.76 [0.42–1.35]; 0.32 | NA | 0.63 [0.38–1.04]; 0.054 | 0.58 [0.36–0.95]; 0.03 | 0.83 [0.42–1.65]; 0.56 | NA |
| Rhinitis | 113 | 41.2 | 21 | 32.3 | 34 | 35.8 | 1.47 [0.80–2.75]; 0.19 | 1.83 [1.01–3.31]; 0.047 | 1.26 [0.76–2.11]; 0.35 | NA | 0.86 [0.41–1.76]; 0.65 | NA |
| Dyspnoea | 79 | 28.8 | 18 | 27.7 | 15 | 15.8 | 1.06 [0.56–2.06]; 0.86 | NA | 2.16 [1.15–4.28]; 0.01 | 2.35 [1.26–4.39]; 0.007 | 2.04 [0.88–4.79]; 0.07 | 2.42 [1.05–5.58]; 0.038 |
| Anosmia | 78 | 28.6 | 14 | 21.5 | 21 | 22.1 | 1.46 [0.74–3.02]; 0.25 | NA | 1.41 [0.79–2.58]; 0.22 | NA | 0.97 [0.41–2.21]; 0.93 | NA |
| Ageusia | 73 | 26.7 | 16 | 24.6 | 20 | 21.1 | 1.12 [0.58–2.24]; 0.73 | NA | 1.37 [0.76–2.54]; 0.27 | NA | 1.22 [0.54–2.76]; 0.60 | NA |
| Hypoxemia | 55 | 20.1 | 7 | 10.8 | 15 | 15.8 | 2.08 [0.88–5.69]; 0.08 | – | 1.34 [0.70–2.70]; 0.36 | NA | 0.64 [0.21–1.81]; 0.37 | NA |
| Clinical outcomes | ||||||||||||
| Hospitalisation | 63 | 23.0 | 15 | 23.1 | 26 | 27.4 | 0.99 [0.51–2.04]; 0.99 | NA | 0.79 [0.45–1.41]; 0.39 | NA | 0.80 [0.35–1.75]; 0.54 | NA |
| ICU | 5 | 1.8 | 1 | 1.5 | 3 | 3.2 | 1.19 [0.13–57.12]; 0.87 | NA | 0.57 [0.11–3.75]; 0.44 | NA | 0.48 [0.01–6.15]; 0.52 | NA |
| Death | 11 | 4.0 | 2 | 3.1 | 3 | 3.2 | 1.32 [0.28–12.52]; 0.72 | NA | 1.28 [0.33–7.31]; 0.71 | NA | 0.97 [0.08–8.75]; 0.98 | NA |
Ref: reference, NA: not applicable, −: non-significant.
In multivariate analysis, patients infected with 20A and 20C viruses were significantly older than those infected with 20B viruses (P-value < 0.01 and <0.05, respectively). Patients infected with the 20A viruses were significantly more likely to present a rhinitis than those infected with the 20B viruses (OR = 1.83, P = 0.047) and were less likely to report a cough than those infected with 20C viruses (OR = 0.58, P = 0.03). Patients infected with 20A and 20B viruses were more likely to present a dyspnoea than patients infected with 20C viruses (OR = 2.35, P = 0.007 and OR = 2.42, P = 0.038, respectively). No significant differences in clinical outcomes, including rates of hospitalisation, transfer to ICU, and death, were observed between patients infected with viruses of these three clades.
3.2. Comparison of patients infected with B.1.177 and B.1.160 variants during the second phase of the epidemic
Table 2 shows the characteristics of 350 patients infected with the two major variants circulating during the second phase of the COVID-19 epidemic in Marseille.
Table 2.
Characteristics of COVID-19 patients infected with Marseille-2 (B.1.177 lineage) and Marseille-4 (B.1.160 lineage) variants during the second phase of the epidemic in Marseille (univariate and multivariate analysis).
| B.1.177 |
B.1.160 |
B.1.177 vs. B.1.160 |
||||
|---|---|---|---|---|---|---|
| n |
% |
n |
% |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
|
| N = 69 | N = 281 | Ref = B.1.160 | ||||
| Sociodemographic characteristic | ||||||
| Mean age | 56.0 ± 21.6 | 58.3 ± 23.1 | NA | NA | ||
| Age | ||||||
| <45 | 24 | 34.8 | 74 | 26.3 | ref | ref |
| 45–64 | 19 | 27.5 | 88 | 31.3 | 0.67 [0.34–1.31]; 0.24 | NA |
| ≥65 | 26 | 37.7 | 119 | 42.4 | 0.67 [0.36–1.26]; 0.22 | NA |
| Gender | ||||||
| Female | 38 | 55.1 | 127 | 45.2 | 0.67 [0.38–1.18]; 0.14 | – |
| Male | 31 | 44.9 | 154 | 54.8 | ||
| Hypertension | 17 | 24.6 | 86 | 30.6 | 0.74 [0.38–1.39]; 0.33 | NA |
| Diabetes | 5 | 7.3 | 49 | 17.4 | 0.37 [0.11–0.98]; 0.04 | – |
| Cancer | 8 | 11.6 | 27 | 9.6 | 1.23 [0.46–2.97]; 0.62 | NA |
| Chronic respiratory disease | 5 | 7.3 | 37 | 13.2 | 0.52 [0.15–1.39]; 0.18 | – |
| Chronic heart disease | 14 | 20.3 | 55 | 19.6 | 1.05 [0.50–2.08]; 0.89 | NA |
| Obesity | 8 | 11.6 | 27 | 9.6 | 1.23 [0.46–2.97]; 0.62 | NA |
| Clinical signs | ||||||
| Fever | 27 | 39.1 | 121 | 43.1 | 0.85 [0.48–1.50]; 0.55 | NA |
| Cough | 28 | 40.6 | 106 | 37.7 | 1.13 [0.63–1.99]; 0.66 | NA |
| Rhinitis | 10 | 14.5 | 45 | 16.0 | 0.89 [0.38–1.92]; 0.76 | NA |
| Dyspnoea | 17 | 24.6 | 71 | 25.3 | 0.97 [0.49–1.83]; 0.91 | NA |
| Anosmia | 11 | 15.9 | 39 | 13.9 | 1.17 [0.51–2.51]; 0.67 | NA |
| Ageusia | 9 | 13.0 | 38 | 13.6 | 0.96 [0.38–2.15]; 0.91 | NA |
| Hypoxemia | 17 | 24.6 | 86 | 30.6 | 0.74 [0.38–1.39]; 0.33 | NA |
| Clinical outcomes | ||||||
| Hospitalisation | 21 | 30.4 | 129 | 45.9 | 0.52 [0.28–0.93]; 0.02 | 0.52 [0.29–0.91]; 0.02 |
| ICU | 2 | 2.9 | 16 | 5.7 | 0.49 [0.05–2.19]; 0.35 | NA |
| Death | 8 | 11.6 | 44 | 15.7 | 0.71 [0.27–1.62]; 0.40 | NA |
Ref: reference, NA: not applicable, −: non-significant.
In multivariate analysis, patients infected with the B.1.177 variant were less likely to be hospitalised than those infected with the B.1.160 variant (OR = 0.52, P = 0.02). No significant differences in demographics, comorbidities, clinical profiles, transfer to the ICU, and mortality were observed.
3.3. Comparison of patients infected with alpha, beta, gamma and A.27 variants during the third phase of the epidemic
Table 3 shows the characteristics of 976 patients infected with the four major variants circulating during the third phase of the COVID-19 epidemic in Marseille.
Table 3.
Characteristics of COVID-19 patients infected with the alpha, beta, gamma and A.27 variants during the third phase of the epidemic in Marseille (univariate analysis).
| Alpha (N = 817) |
Beta (N = 98) |
Gamma (N = 20) |
A.27 (N = 41) |
Alpha vs. beta |
Gamma vs. alpha |
Gamma vs. beta |
A.27 vs. alpha |
A.27 vs. beta |
A.27 vs. gamma |
|
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) |
n (%) |
n (%) |
n (%) |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
|
| Ref = beta | Ref = alpha | Ref = beta | Ref = alpha | Ref = beta | Ref = gamma | |||||
| Sociodemographic characteristic | ||||||||||
| Mean age | 54.6 ± 17.5 | 52.9 ± 19.0 | 52.6 ± 13.7 | 56.0 ± 19.2 | NA | NA | NA | NA | NA | NA |
| Age | ||||||||||
| <45 | 229 (28.0) | 29 (29.6) | 3 (15.0) | 12 (29.3) | ref | ref | ref | ref | ref | ref |
| 45–64 | 340 (41.6) | 39 (39.8) | 15 (75.0) | 15 (36.6) | 1.10 [0.66–1.84]; 0.70 | 3.37 [0.96–11.76]; 0.06 | 3.72 [0.98–14.05]; 0.053 | 0.84 [0.39–1.83]; 0.66 | 0.93 [0.38–2.28]; 0.87 | 0.25 [0.06–1.07]; 0.06 |
| ≥65 | 248 (30.4) | 30 (30.6) | 2 (10.0) | 14 (34.1) | 1.05 [0.61–1.80]; 0.87 | 0.62 [0.10–3.72]; 0.60 | 0.64 [0.10–4.14]; 0.64 | 1.08 [0.49–2.38]; 0.85 | 1.13 [0.45–2.84]; 0.80 | 1.75 [0.25–12.28]; 0.57 |
| Gender | ||||||||||
| Female | 404 (49.5) | 48 (49.0) | 8 (40.0) | 22 (53.7) | ref | ref | ref | ref | ref | ref |
| Male | 413 (50.5) | 50 (51.0) | 12 (60.0) | 19 (46.3) | 0.98 [0.63–1.53]; 0.93 | 1.47 [0.54–4.18]; 0.40 | 1.44 [0.49–4.43]; 0.46 | 0.84 [0.43–1.66]; 0.60 | 0.83 [0.37–1.83]; 0.61 | 0.58 [0.17–1.93]; 0.32 |
| Hypertension | 226 (27.7) | 30 (30.6) | 5 (25.0) | 11 (26.8) | 0.87 [0.54–1.42]; 0.54 | 0.87 [0.25–2.56]; 0.79 | 0.76 [0.20–2.46]; 0.62 | 0.96 [0.43–2.01]; 0.91 | 0.83 [0.33–2.00]; 0.66 | 1.10 [0.28–4.79]; 0.88 |
| Diabetes | 109 (13.3) | 10 (10.2) | 2 (10.0) | 5 (12.2) | 1.35 [0.68–3.01]; 0.38 | 0.72 [0.08–3.08]; 0.66 | 0.98 [0.10–5.20]; 0.98 | 0.90 [0.27–2.38]; 0.83 | 1.22 [0.31–4.26]; 0.73 | 1.25 [0.18–14.29]; 0.80 |
| Cancer | 54 (6.6) | 6 (6.1) | 0 (0) | 4 (9.8) | 1.09 [0.45–3.17]; 0.85 | NA | NA | 1.53 [0.38–4.49]; 0.43 | 1.66 [0.32–7.43]; 0.45 | NA |
| Chronic respiratory disease | 100 (12.2) | 12 (12.2) | 2 (10.0) | 1 (2.4) | 1.00 [0.52–2.08]; 1.00 | 0.80 [0.09–3.41]; 0.76 | 0.80 [0.08–4.08]; 0.78 | 0.18 [0.01–1.08]; 0.06 | 0.18 [0.01–1.30]; 0.07 | 0.23 [0.01–4.70]; 0.19 |
| Chronic heart disease | 72 (8.8) | 10 (10.2) | 0 (0) | 2 (4.9) | 0.85 [0.42–1.92]; 0.65 | NA | NA | 0.53 [0.06–2.13]; 0.38 | 0.45 [0.05–2.28]; 0.31 | NA |
| Obesity | 162 (19.8) | 13 (13.3) | 5 (25.0) | 8 (19.5) | 1.62 [0.87–3.24]; 0.12 | 1.35 [0.38–2.78]; 0.94 | 2.18 [0.53–7.76]; 0.18 | 0.98 [0.38–2.22]; 0.96 | 0.16 [0.52–4.57]; 0.35 | 0.73 [0.17–3.34]; 0.62 |
| Clinical signs | ||||||||||
| Fever | 361 (44.2) | 34 (34.7) | 9 (45.0) | 13 (31.7) | 1.49 [0.94–2.38]; 0.07 | 1.03 [0.37–2.78]; 0.94 | 1.54 [0.51–4.53]; 0.38 | 0.59 [0.27–1.19]; 0.12 | 0.87 [0.37–2.02]; 0.73 | 0.57 [0.17–1.98]; 0.31 |
| Cough | 384 (47.0) | 38 (38.8) | 13 (65.0) | 16 (39.0) | 1.40 [0.89–2.21]; 0.12 | 2.09 [0.77–6.26]; 0.11 | 2.93 [0.97–9.42]; 0.03 | 0.72 [0.35–1.43]; 0.32 | 1.01 [0.44–2.27]; 0.98 | 0.34 [0.10–1.19]; 0.06 |
| Rhinitis | 176 (21.5) | 16 (16.3) | 3 (15.0) | 10 (24.4) | 1.41 [0.79–2.64]; 0.23 | 0.64 [0.12–2.26]; 0.48 | 0.90 [0.15–3.70]; 0.88 | 1.17 [0.50–2.52]; 0.67 | 1.65 [0.60–4.36]; 0.27 | 1.83 [0.39–11.62]; 0.40 |
| Dyspnoea | 211 (25.8) | 25 (25.5) | 4 (20.0) | 7 (17.1) | 1.02 [0.62–1.72]; 0.95 | 0.72 [0.17–2.26]; 0.56 | 0.73 [0.16–2.57]; 0.60 | 0.59 [0.22–1.38]; 0.21 | 0.60 [0.20–1.62]; 0.28 | 0.82 [0.18–4.42]; 0.78 |
| Anosmia | 91 (11.1) | 15 (15.5) | 7 (35.0) | 2 (4.9) | 0.69 [0.37–1.34]; 0.21 | 4.30 [1.41–11.91]; 0.001 | 2.94 [0.84–9.53]; 0.04 | 0.41 [0.05–1.63]; 0.21 | 0.28 [0.03–1.31]; 0.08 | 0.10 [0.01–0.61]; 0.002 |
| Ageusia | 83 (10.2) | 12 (12.4) | 5 (25.0) | 2 (4.9) | 0.80 [0.41–1.68]; 0.50 | 2.95 [0.82–8.80]; 0.03 | 2.36 [0.56–8.54]; 0.14 | 0.45 [0.05–1.81]; 0.27 | 0.36 [0.04–1.76]; 0.18 | 0.15 [0.01–1.10]; 0.02 |
| Hypoxemia | 199 (24.4) | 30 (30.6) | 3 (15.0) | 11 (26.8) | 0.73 [0.45–1.20]; 0.18 | 0.55 [0.10–1.92]; 0.33 | 0.40 [0.07–1.55]; 0.16 | 1.14 [0.51–2.39]; 0.72 | 0.83 [0.33–2.00]; 0.66 | 2.08 [0.45–13.05]; 0.30 |
| Clinical outcomes | ||||||||||
| Hospitalisation | 203 (24.9) | 31 (31.6) | 4 (20.0) | 11 (26.8) | 0.71 [0.45–1.17]; 0.15 | 0.76 [0.18–2.38]; 0.62 | 0.54 [0.12–1.88]; 0.30 | 1.11 [0.49–2.33]; 0.77 | 0.79 [0.32–1.89]; 0.57 | 1.47 [0.35–7.31]; 0.56 |
| ICU | 58 (7.1) | 9 (9.2) | 2 (10.0) | 2 (4.9) | 0.76 [0.36–1.80]; 0.45 | 1.45 [0.16–6.31]; 0.62 | 1.10 [0.11–5.98]; 0.91 | 0.67 [0.08–2.71]; 0.59 | 0.51 [0.05–2.63]; 0.39 | 0.46 [0.03–6.93]; 0.45 |
| Death | 34 (4.2) | 0 (0) | 0 (0) | 4 (9.8) | NA | NA | NA | 2.49 [0.61–7.52]; 0.09 | NA | NA |
SdA: South African, Ref: reference, NA: not applicable.
Patients infected with alpha and beta variants presented a relatively similar profile of demographic characteristics, comorbidities, and clinical symptoms in multivariate analysis (Table 4 ). Patients infected with the gamma variant were more frequently in the 45–64 years of age group than those infected with the alpha (OR = 4.21, P = 0.006), beta (OR = 4.89, P = 0.005) or A.27 (OR = 4.17, P = 0.03) variants. Patients infected with the gamma variant were more likely to present with a cough than those infected with the beta variant (OR = 3.24, P = 0.028) and were more likely to report anosmia than those infected with the alpha (OR = 4.30, P = 0.003) and A.27 (OR = 8.33, P = 0.02) variants. No significant differences of clinical outcomes (hospitalisation, transfer to the ICU and death) were observed between patients infected with these four variants.
Table 4.
Characteristics of COVID-19 patients infected with the alpha, beta, gamma and A-27 (Marseille-501) variants during the third phase of the epidemic in Marseille (multivariate analysis).
| Alpha vs. beta |
Gamma vs. alpha |
Gamma vs. beta |
A.27 vs. alpha |
A.27 vs. beta |
A.27 vs. gamma |
|
|---|---|---|---|---|---|---|
| OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
OR [95% CI]; P-value |
|
| Ref = beta | Ref = alpha | Ref = beta | Ref = alpha | Ref = beta | Ref = gamma | |
| Sociodemographic characteristic | ||||||
| Age | ||||||
| <45 | NA | ref | ref | ref | ref | ref |
| 45–64 | NA | 4.21 [1.51–11.77]; 0.006 | 4.89 [1.59–14.98]; 0.005 | NA | NA | 0.24 [0.07–0.83]; 0.03 |
| ≥65 | NA | – | NA | NA | NA | – |
| Gender | ||||||
| Female | ||||||
| Male | NA | NA | NA | NA | NA | NA |
| Hypertension | NA | NA | NA | NA | NA | NA |
| Diabetes | NA | NA | NA | NA | NA | NA |
| Cancer | NA | NA | NA | NA | NA | NA |
| Chronic respiratory disease | NA | NA | NA | – | – | – |
| Chronic heart disease | NA | NA | NA | NA | NA | NA |
| Obesity | – | NA | – | NA | NA | NA |
| Clinical signs | ||||||
| Fever | – | NA | NA | – | NA | NA |
| Cough | – | – | 3.24 [1.13–9.28]; 0.028 | NA | NA | – |
| Rhinitis | NA | NA | NA | NA | NA | NA |
| Dyspnoea | NA | NA | NA | NA | NA | NA |
| Anosmia | NA | 4.30 [1.66–11.19]; 0.003 | – | NA | – | 0.12 [0.02–0.73]; 0.02 |
| Ageusia | NA | – | – | NA | – | – |
| Hypoxemia | – | NA | – | NA | NA | NA |
| Clinical outcomes | ||||||
| Hospitalisation | – | NA | NA | NA | NA | NA |
| ICU | NA | NA | NA | NA | NA | NA |
| Death | NA | NA | NA | – | NA | NA |
SdA: South African, Ref: reference, NA: not applicable, −: non-significant.
3.4. Comparison of COVID-19 patients during three phases of the epidemic
Table 5 shows the characteristics of 1760 patients infected during the three phases of the epidemic in Marseille.
Table 5.
Characteristics of COVID-19 patients during the three phases of the epidemic in Marseille (univariate and multivariate analysis).
| First phase |
Second phase |
Third phase |
Second phase vs. first phase |
Third phase vs. first phase |
Second phase vs. third phase |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N = 434 |
N = 350 |
N = 976 |
||||||||||
| n |
% |
n |
% |
n |
% |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
Univariate analysis OR [95% CI]; P-value |
Multivariate analysis OR [95% CI]; P-value |
|
| N = 350 | Ref = first phase | Ref = first phase | Ref = third phase | |||||||||
| Sociodemographic characteristic | ||||||||||||
| Mean age | 47.5 ± 19.5 | 57.8 ± 22.8 | 54.4 ± 17.7 | NA | NA | NA | NA | NA | NA | |||
| Age < 45 | 204 | 47.0 | 98 | 28.0 | 273 | 28.0 | ref | ref | ref | ref | ref | ref |
| 45–64 | 151 | 34.8 | 107 | 30.6 | 409 | 41.9 | 1.48 [1.04–2.08]; 0.03 | – | 2.02 [1.56–2.63]; <0.0001 | 1.75 [1.32–2.30]; <0.0001 | 0.73 [0.53–0.99]; 0.048 | – |
| ≥65 | 79 | 18.2 | 145 | 41.4 | 294 | 30.1 | 3.82 [2.65–5.50]; <0.0001 | 2.20 [1.54–3.14]; <0.0001 | 2.78 [2.04–3.78]; <0.0001 | 2.45 [1.71–3.49]; <0.0001 | 1.37 [1.01–1.86]; 0.04 | – |
| Gender | ||||||||||||
| Female | 234 | 53.9 | 165 | 47.1 | 482 | 49.4 | Ref | Ref | Ref | Ref | Ref | Ref |
| Male | 200 | 46.1 | 185 | 52.9 | 494 | 50.6 | 1.31 [0.98–1.76]; 0.06 | – | 1.20 [0.95–1.51]; 0.12 | – | 1.09 [0.85–1.41]; 0.47 | NA |
| Hypertension | 79 | 18.2 | 103 | 29.4 | 272 | 27.9 | 1.87 [1.32–2.66]; 0.0002 | – | 1.74 [1.30–2.33]; 0.0001 | – | 1.08 [0.82–1.42]; 0.58 | NA |
| Diabetes | 36 | 8.3 | 54 | 15.4 | 126 | 12.9 | 2.01 [1.17–3.25]; 0.002 | – | 1.64 [1.10–2.49]; 0.01 | – | 1.23 [0.85–1.76]; 0.24 | NA |
| Cancer | 16 | 3.7 | 35 | 10.0 | 64 | 6.6 | 2.90 [1.53–5.71]; 0.0004 | – | 1.83 [1.03–3.44]; 0.03 | – | 1.58 [0.99–2.48]; 0.04 | – |
| Chronic respiratory disease | 44 | 10.1 | 42 | 12.0 | 115 | 11.8 | 1.21 [0.75–1.94]; 0.41 | NA | 1.18 [0.81–1.75]; 0.37 | NA | 1.02 [0.68–1.50]; 0.91 | NA |
| Chronic heart disease | 47 | 10.8 | 69 | 19.7 | 84 | 8.6 | 2.02 [1.33–3.09]; 0.0005 | – | 0.78 [0.52–1.16]; 0.18 | 0.40 [0.26–0.61]; <0.0001 | 2.61 [1.81–3.73]; <0.0001 | 1.83 [1.23–2.70]; 0.003 |
| Obesity | 46 | 10.6 | 35 | 10.0 | 188 | 19.3 | 0.94 [0.57–1.53]; 0.78 | NA | 2.01 [1.41–2.91]; 0.0001 | 1.88 [1.31–2.70]; 0.001 | 0.47 [0.31–0.69]; 0.0001 | 0.45 [0.30–0.67]; <0.0001 |
| Clinical signs | ||||||||||||
| Fever | 118 | 27.2 | 148 | 42.3 | 417 | 42.7 | 1.96 [1.44–2.68]; <0.0001 | 2.27 [1.60–3.11]; <0.0001 | 2.00 [1.55–2.58]; <0.0001 | 1.94 [1.50–2.52]; <0.0001 | 0.98 [0.76–1.27]; 0.89 | NA |
| Cough | 226 | 52.1 | 134 | 38.3 | 451 | 46.2 | 0.57 [0.42–0.77]; 0.0001 | 0.69 [0.50–0.96]; 0.03 | 0.79 [0.63–0.99]; 0.04 | – | 0.72 [0.56–0.93]; 0.01 | – |
| Rhinitis | 168 | 38.7 | 55 | 15.7 | 205 | 21.0 | 0.30 [0.20–0.42]; <0.0001 | 0.37 [0.26–0.54]; <0.0001 | 0.42 [0.33–0.54]; <0.0001 | 0.51 [0.39–0.67]; <0.0001 | 0.70 [0.50–0.98]; 0.03 | – |
| Dyspnoea | 112 | 25.8 | 88 | 25.1 | 247 | 25.3 | 0.97 [0.69–1.35]; 0.83 | NA | 0.97 [0.75–1.27]; 0.84 | NA | 0.99 [0.74–1.32]; 0.95 | NA |
| Anosmia | 113 | 26.1 | 50 | 14.3 | 115 | 11.8 | 0.47 [0.32–0.69]; 0.0001 | – | 0.38 [0.28–0.51]; <0.0001 | – | 1.25 [0.86–1.81]; 0.22 | NA |
| Ageusia | 109 | 25.2 | 47 | 13.5 | 102 | 10.5 | 0.46 [0.31–0.68]; <0.0001 | – | 0.35 [0.25–0.47]; <0.0001 | 0.46 [0.34–0.64]; <0.0001 | 1.33 [0.90–1.95]; 0.13 | 1.73 [1.18–2.54]; 0.005 |
| Hypoxemia | 77 | 17.7 | 103 | 29.4 | 243 | 24.9 | 1.93 [1.36–2.75]; 0.0001 | – | 1.54 [1.15–2.07]; 0.003 | – | 1.26 [0.95–1.66]; 0.10 | – |
| Clinical outcomes | ||||||||||||
| Hospitalisation | 104 | 24.0 | 150 | 42.9 | 249 | 25.5 | 2.38 [1.73–3.27]; <0.0001 | – | 1.09 [0.83–1.43]; 0.54 | NA | 2.19 [1.68–2.85]; <0.0001 | 1.98 [1.43–2.74]; <0.0001 |
| ICU | 9 | 2.1 | 18 | 5.1 | 71 | 7.3 | 2.56 [1.07–6.55]; 0.02 | – | 3.70 [1.82–8.51]; 0.0001 | 2.30 [1.11–4.78]; 0.03 | 0.69 [0.38–1.19]; 0.17 | 0.32 [0.18–0.58]; <0.0001 |
| Death | 16 | 3.7 | 52 | 14.9 | 38 | 3.9 | 4.56 [2.50–8.71]; <0.0001 | 1.91 [1.02–3.59]; 0.04 | 1.06 [0.57–2.06]; 0.85 | NA | 4.31 [2.72–6.86]; <0.0001 | 3.01 [1.81–5.01]; <0.0001 |
Ref: reference, NA: not applicable, −: non significant.
In multivariate analysis, compared to patients in the first phase, those in the second phase were older (OR = 2.20, P < 0.0001) and more likely to present with a fever (OR = 2.27, P < 0.0001), but less likely to report a cough (OR = 0.69, P = 0.03) and rhinitis (OR = 0.37, P < 0.0001). Patients in the second phase were also more likely to die than those in the first phase (OR = 1.91, P = 0.04). Compared to patients in the first phase, patients infected during the third phase were significantly older and less likely to report chronic heart diseases (OR = 0.40, P < 0.0001) but more likely to report obesity (OR = 1.88, P = 0.001). They were more likely to present with a fever (OR = 1.94, P < 0.0001), but less likely to report rhinitis and ageusia (OR = 0.51, P < 0.0001 and OR = 0.46, P < 0.0001, respectively). Finally, they were more frequently transferred to an ICU than those in the first phase (OR = 2.30, P = 0.03). Compared to patients in the third phase, patients infected during the second phase were more likely to report chronic heart diseases (OR = 1.83, P = 0.003) but less likely to report obesity (OR = 0.45, P < 0.0001). They were more likely to report ageusia (OR = 1.73, P = 0.005). Finally, they were more likely to be hospitalised and to die (OR = 1.98, P < 0.0001 and OR = 3.01, P < 0.0001, respectively), but were less likely to be transferred to an ICU (OR = 0.32, P < 0.0001).
4. Discussion
SARS-CoV-2 is able to rapidly genetically diversify and can spread internationally through travellers and cause successive outbreaks. SARS-CoV-2 variants cause the COVID-19 disease with different clinical manifestations, even in populations who have previously been exposed to original viruses (Zeyaullah et al., 2021; SeyedAlinaghi et al., 2021; Gautret et al., 2021). In this study, we compared the demographics, clinical profile, and outcomes of patients infected with different variants that circulated or continue to circulate in the Marseille area, where three major phases have occurred until June 2021. During each phase, no major differences were observed with regards to age and gender distribution, the prevalence of chronic diseases, and clinical symptoms between variants circulating during a given phase. The overall comparison between the three phases from March 2020 to April 2021 showed that the variants of the second phase (B.1.177 (Marseille-2) and B.1.160 (Marseille-4) lineages, respectively) were associated with more severe outcomes, leading to higher rates of hospitalisation and death. The appearance of the variants of concern harbouring the N501Y substitution at the end of December 2020 caught the attention of the World Health Organization because of faster spreading than other variants and the original viruses (European Centre for Disease Prevention and Control, 2021; PAHO, 2021; Lauring and Hodcroft, 2021; Frampton et al., 2021). However, the association between these variants and severity of COVID-19 disease remains unclear (Dao et al., 2021b). In studies that analysed community-based testing datasets, the alpha variant was significantly associated with an increase in hospitalisation and death (Challen et al., 2021; Davies et al., 2021; Grint et al., 2021). In contrast, in a clinical study, Frampton et al. showed that this variant was not associated with the severity of COVID-19 disease (Frampton et al., 2021). In addition, an analysis of UK COVID-19 Clinical Information Network data showed no higher risk of in-hospital deaths in patients infected with the alpha variant (GOVUK, 2021). There was also no evidence of an increased risk for hospitalisation among patients infected with this variant (GOVUK, 2021). Our observations corroborate other clinical studies. Patients infected during the third phase with variants harbouring the N501Y substitution were more frequently obese than others. We have no explanation for this association, and it is probably a random event.
Monitoring the epidemiology of SARS-CoV-2 variants is very important for epidemic control. As of 11 May 2021, 46,251 replacements in SARS-CoV-2 proteins were identified according to the CoV-GLUE online tool (http://cov-glue.cvr.gla.ac.uk/#/replacement). The highest numbers of these amino acid replacements are related to the NSP3 and S proteins, in 9414 (20.4%) and 6238 (13.5%) cases, respectively. Recently, the B1.617 variant of SARS-CoV-2 emerged in Maharashtra, India and has since then spread to at least 17 countries (https://www.gisaid.org/). At the time of writing this manuscript, only a few cases had been documented in Marseille (Scola et al., 2021). The presentation and severity of the disease may be very different depending on the SARS-CoV-2 variants, so the public health response needs to be adapted in real-time to the genomic profile of each viral epidemic phase.
This work has some limitations. We did not evaluate the transmissibility of different variants. The number of patients infected with different variants analysed in this work ranging from 20 to 817 may indirectly indicate the different degree of spread of the variants. In addition, we only analysed patients seen at our hospitals (IHU and AP-HM wards), requiring medical care with a viral load allowing successful virus genome sequencing. This could introduce a major selection bias. A large proportion of patients were excluded from our analysis because they were not treated either at IHUor at AP-HM, and the actual severity of SARS-CoV-2 variants may differ from our results. The patients who were not included here were significantly younger and could have been asymptomatic or pauci-symptomatic. This selection bias is notably evidenced by the significantly higher hospitalisation, transfer to ICU and mortality rates observed in patients whose virus sequence was available, as compared to those of the overall population of COVID-19 patients seen at our Institute with rates of hospitalisation of about 18%, of transfer to ICU of about 2%, and with mortality of about 1% (Lagier et al., 2020). Moreover, access to care may vary according to the phases of the epidemic and potentially influence the severity of the disease. In addition, several biomarkers known to be associated with severity, including thrombopenia, D-dimer counts, troponin level and lactate dehydrogenase (Henry et al., 2020), were not considered in this analysis. Furthermore, we did not provide information on the duration of symptoms which could differ by variant.
Nevertheless, our study is the largest clinical study to date that compared the clinical profiles of nine lineages of SARS-CoV-2. A large cohort study is recommended to evaluate the transmissibility and better characterise the clinical severity of emerging variants.
The following are the supplementary data related to this article.
Age and gender in patients with available clinical data and in those excluded from the study.
Funding
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the French National Research Agency under the “Investissements d'avenir” programme, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d'Azur and European FEDER PRIMI funding.
Credit author statement
Conceptualization: PG, DR, VTH; Data curation: VTH, PC, AL, JD, JCL, PP, PEF and MM ; Formal analysis: VTH, PC, AL, JD; Investigation: VTH, PC, AL, DR, PG; Methodology: VTH, PC, AL, JD, PG; Project administration: DR, PG; Resources: PC, AL, JD, JCL, PP, PEF and MM; Software: VTH, PC, AL, JD; Supervision: DR, PG; Validation: VTH, PC, AL, JD, JCL, PP, PEF, MM, DR, PG; Visualization: PC, DR, PG; Writing - original draft: VTH; Writing - review & editing: PC, AL, JD, JCL, PP, PEF, MM, DR, PG.
Declaration of Competing Interest
VTH, PC, AL, JD, JCL, PP, MM, PEF, DR and PG declare that they have no conflicts of interest.
Acknowledgments
Our thanks go to Ludivine Brechard, Vera Esteves-Vieira, Elsa Prudent, Marielle Bedotto, Linda Houhamdi and all the staff at the IHU Méditerranée Infection for their support with molecular techniques.
References
- Amrane S., Tissot-Dupont H., Doudier B., Eldin C., Hocquart M., Mailhe M., et al. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, − January 31st to March 1st, 2020: a respiratory virus snapshot. Travel Med. Infect. Dis. 2020;36:101632. doi: 10.1016/j.tmaid.2020.101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedotto M., Fournier P.E., Houhamdi L., Colson P., Raoult D. Implementation of an in-house real-time reverse transcription-PCR assay to detect the emerging SARS-CoV-2 N501Y variants. J. Clin. Virol. 2021;140:104868. doi: 10.1016/j.jcv.2021.104868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac Z., Gauss C.H., Williams D.K., Hosmer D.W. Purposeful selection of variables in logistic regression. Source Code Biol. Med. 2008;3:17. doi: 10.1186/1751-0473-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The mutation that helps Delta spread like wildfire. Nature. 2021;596(7873):472–473. doi: 10.1038/d41586-021-02275-2. [DOI] [PubMed] [Google Scholar]
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Lagier J.C., Baudoin J.P., Bou Khalil J., La Scola B., Raoult D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(8):1601–1603. doi: 10.1007/s10096-020-03869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Levasseur A., Gautret P., Fenollar F., Thuan Hoang V., Delerce J., et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: an investigational study. Travel Med. Infect. Dis. 2021;27(9):1352.e1–1352.e5. doi: 10.1016/j.cmi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Fournier P.E., Chaudet H., Delerce J., Giraud-Gatineau A., Houhamdi L., Andrieu C., Brechard L., Bedotto M., Prudent E., Gazin C., Beye M., Burel E., Dudouet P., Tissot-Dupont H., Gautret P., Lagier J.C., Million M., Brouqui P., Parola P., Drancourt M., La Scola B., Levasseur A., Raoult D. Analysis of SARS-CoV-2 variants from 24,181 patients exemplifies the role of globalisation and zoonosis in pandemics. medRxiv. 2021 doi: 10.1101/2021.09.10.21262922. [Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Levasseur A., Delerce J., Pinaul L., Dudouet P., Devaux C., Fournier P.E., La Scola B., Lagier J.C., Raoult D. Spreading of a new SARS-CoV-2 N501Y spike variant in a new lineage. Clin. Microbiol. Infect. 2021;27(9):1352.e1–1352.e5. doi: 10.1016/j.cmi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.L., Hoang V.T., Nguyen N.N., Delerce J., Chaudet H., Levasseur A., et al. Clinical outcomes in COVID-19 patients infected with different SARS-CoV-2 variants in Marseille, France. Clin. Microbiol. Infect. 2021;24 doi: 10.1016/j.cmi.2021.05.029. S1198-743X(21)00270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.L., Hoang V.T., Colson P., Lagier J.C., Million M., Raoult D., Levasseur A., Gautret P. SARS-CoV-2 infectivity and severity of COVID-19 according to SARS-CoV-2 variants: current evidence. J. Clin. Med. 2021 Jun 15;10(12) doi: 10.3390/jcm10122635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group, Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . European Centre for Disease Prevention and Control; 2021. Risk Assessment: Risk Related to the Spread of New SARS-CoV-2 Variants of Concern in the EU/EEA – First Update.https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update (accessed May 11, 2021) [Google Scholar]
- European Centre for Disease Prevention and Control SARS-CoV-2 Variants of Concern as of 26 August 2021. https://www.ecdc.europa.eu/en/covid-19/variants-concern Available at: (accessed September 01, 2021)
- Faria N.R., Claro I.M., Candido D., Moyses Franco L.A., Andrade P.S., Coletti T.M., et al. Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings. 2021. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 Available at:
- Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P.E., Colson P., Levasseur A., Gautret P., Bedotto M., Filosa V., et al. Genome Sequence Analysis Enabled Deciphering the Atypical Evolution of COVID-19 Epidemics in Marseille, France. 2021. https://www.mediterranee-infection.com/wp-content/uploads/2020/04/Capture-d%E2%80%99%C3%A9cran-2021-02-02-%C3%A0-09.48.22.pdf Available at:
- Fournier P.-E., Colson P., Levasseur A., Devaux C.A., Gautret P., Bedotto M., et al. Emergence and outcomes of the SARS-CoV-2 “Marseille-4” variant. Int. J. Infect. Dis. 2021;106:228–236. doi: 10.1016/j.ijid.2021.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Million M., Jarrot P.-A., Camoin-Jau L., Colson P., Fenollar F., et al. Natural history of COVID-19 and therapeutic options. Expert. Rev. Clin. Immunol. 2020;16:1159–1184. doi: 10.1080/1744666X.2021.1847640. [DOI] [PubMed] [Google Scholar]
- Gautret P., Houhamdi L., Nguyen N.N., Hoang V.T., Giraud-Gatineau A., Raoult D. Does SARS-CoV-2 re-infection depend on virus variant? Clin. Microbiol. Infect. 2021 Jun 28;27(9):1374–1375. doi: 10.1016/j.cmi.2021.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOVUK NERVTAG paper on COVID-19 variant of concern B.1.1.7. 2021. https://www.gov.uk/government/publications/nervtag-paper-on-covid-19-variant-of-concern-b117 (accessed May 11, 2021)
- Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro. Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- Institut National de la Statistique et des Etudes Economiques (INSEE) Fichier des personnes décédées. 2021. https://www.data.gouv.fr/fr/datasets/fichier-des-personnes-decedees/ Available at:
- Lagier J.C., Million M., Gautret P., Colson P., Cortaredona S., Giraud-Gatineau A., et al. Outcomes of 3,737 COVID-19 patients treated with hydroxychloroquine/azithromycin and other regimens in Marseille, France: A retrospective analysis. Travel Med. Infect. Dis. 2020 Jul-Aug;36:101791. doi: 10.1016/j.tmaid.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von HA, et al. IQTREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAHO Epidemiological Update: Variants of SARS-CoV-2 in the Americas - 24 March 2021 - PAHO/WHO | Pan American Health Organization. 2021. https://www.paho.org/en/documents/epidemiological-update-variants-sars-cov-2-americas-24-march-2021 (accessed May 11, 2021)
- La Scola B., Lavrard P., Fournier P.-E., Colson P., Lacoste A., Raoult D. SARS-CoV-2 variant from India to Marseille: The still active role of ports in the introduction of epidemics. Travel Med Infect Dis. 2021;42:102085. doi: 10.1016/j.tmaid.2021.102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SeyedAlinaghi S., Mirzapour P., Dadras O., Pashaei Z., Karimi A., MohsseniPour M., et al. Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review. Eur. J. Med. Res. 2021;26(1):51. doi: 10.1186/s40001-021-00524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- WHO COVID-19 severity | WHO Western Pacific. 2021. https://www.who.int/westernpacific/emergencies/covid-19/information/severity (accessed May 11, 2021)
- Worldometers COVID Live Update: 174,115,762 Cases and 3,745,384 Deaths from the Coronavirus - Worldometer. 2021. https://www.worldometers.info/coronavirus/ (accessed June 07, 2021)
- Zeyaullah M., AlShahrani A.M., Muzammil K., Ahmad I., Alam S., Khan W.H., et al. COVID-19 and SARS-CoV-2 variants: current challenges and health concern. Front. Genet. 2021;12:693916. doi: 10.3389/fgene.2021.693916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Age and gender in patients with available clinical data and in those excluded from the study.