Figure 4. Inositol decline upon IMPA1 loss induces mitochondrial fission in an AMPK-dependent manner.
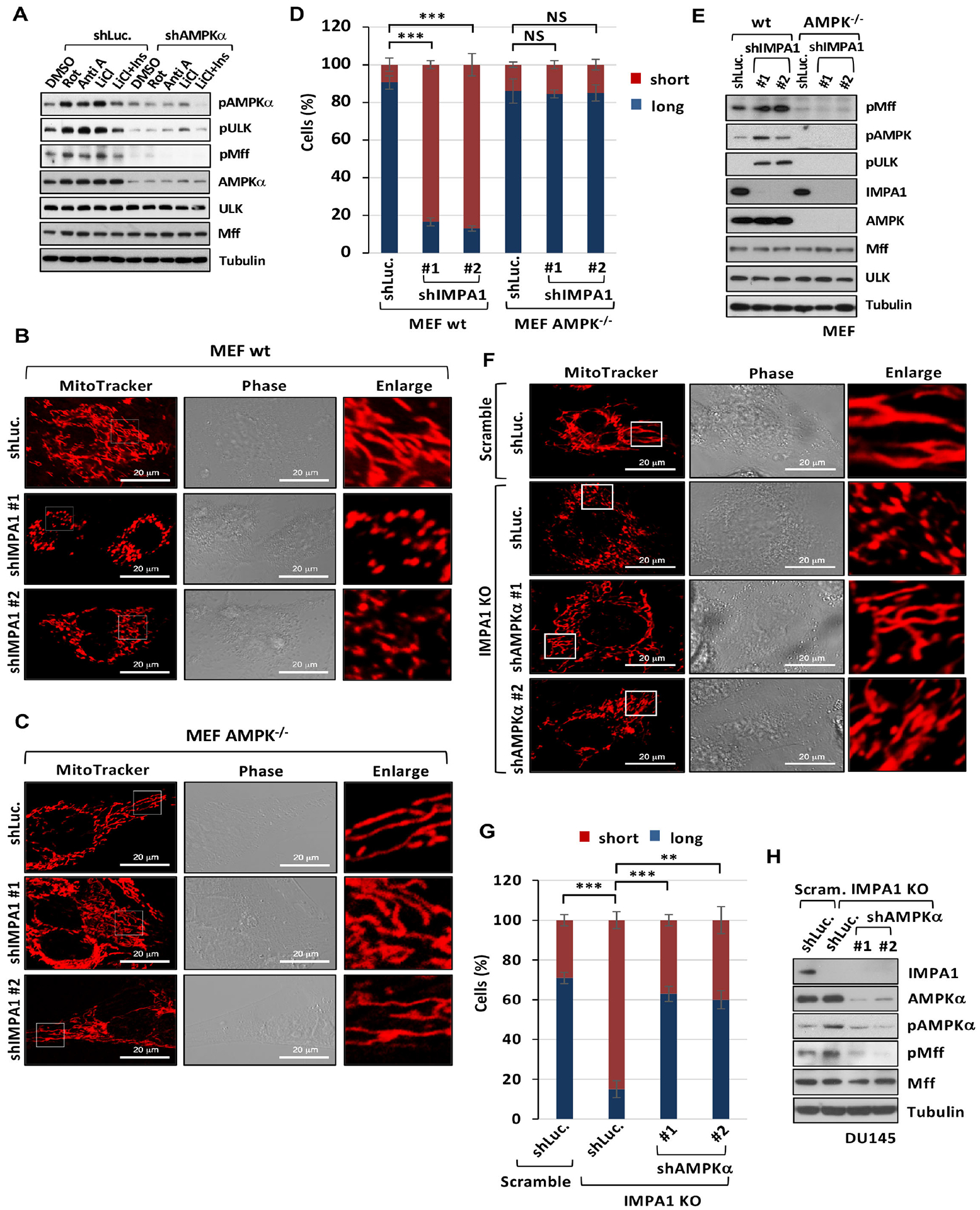
(A) Immunoblotting of shLuc. or shAMPKα DU145 cells treated with DMSO, 250 ng/ml of rotenone (Rot), 10 μM of antimycin A (Anti A), 500 μM of LiCl or 500 μM of LiCl plus 25 μM of Ins. (B and C) Confocal images of mitochondrial morphology in wild type (wt) MEFs and AMPK−/− MEFs stably transduced with shLuc. or shIMPA1. Scale bar, 20 μm. (D) Quantification of the mitochondrial morphology of MEFs shown in (B and C). (E) Immunoblotting of wt and AMPK−/− MEFs stably expressing shLuc. or shIMPA1. (F) Confocal images in CRISPR/Cas9 knockout DU145 cells stably transduced with the guide RNA pairs of scramble (Scram.), IMPA1 knockout clone #1 (IMPA1 KO), shLuc., or shAMPKα. Scale bar, 20 μm. (G) Quantification of the mitochondrial morphology of the DU145 shown in (F). (H) Immunoblotting of CRISPR/Cas9 knockout DU145 cells transduced with the guide RNA pairs of Scram., IMPA1 KO, shLuc., or shAMPKα.
