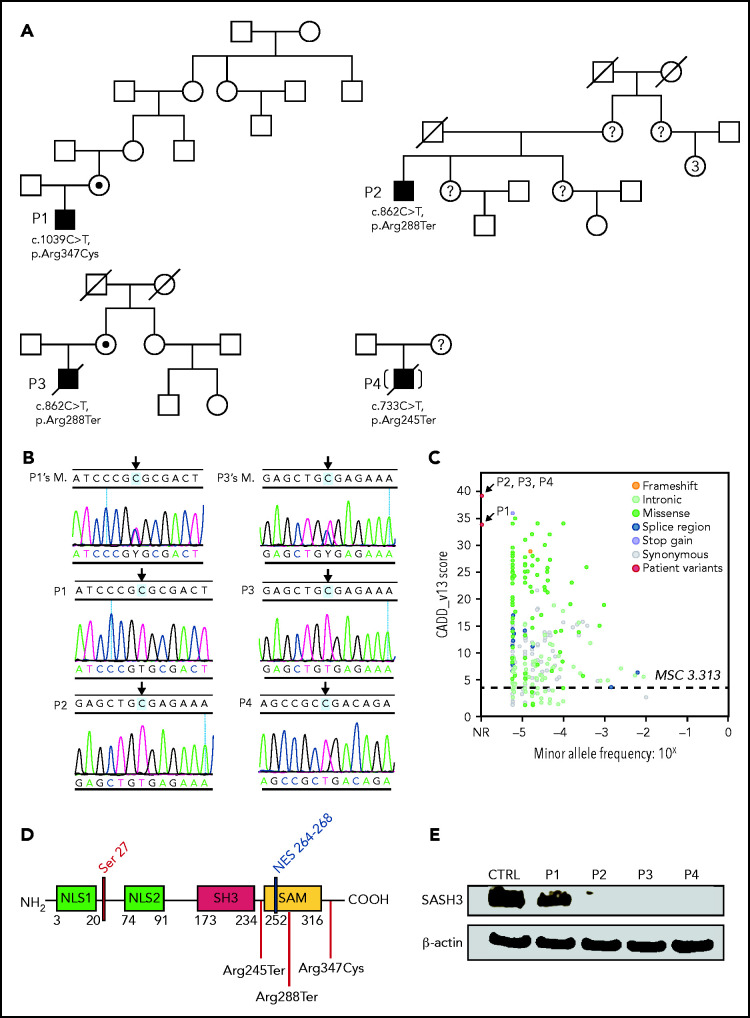
Figure 1.
Pedigrees, SASH3 genetic variants, and protein expression. (A) Pedigrees and familial segregation of mutant SASH3 alleles. Probands are indicated as P1to P4. SASH3 variants detected by whole-exome sequencing are designated at the nucleotide and amino acid level below each patient. (B) Sanger sequencing confirmation of SASH3 variants. The patient is identified on the upper left of each chromatogram (M., mother). Arrows designate the mutated nucleotide. Parental sequencing was not obtained for P2 or for P4 who was adopted. (C) SASH3 variants are predicted to be damaging. Plot of combined annotation depletion dependent (CADD, version 13) score vs minor allele frequency (MAF) of SASH3 modified from PopViz (Rockefeller University, New York, NY). The dotted horizontal line corresponds to the SASH3 mutation significance cutoff (MSC) score. Arrows identify the CADD score of each of the SASH3 variants detected in the patients, all of which were private (MAF = 0). (D) Schematic representation of the SASH3 protein and its domains. Locations of the patients’ variants are indicated by vertical red lines. Numbers indicate amino acid positions. (E) SASH3 protein is not detectable in patients with nonsense SASH3 variants. The immunoblot shows results for SASH3 and β-actin protein expression in PBMC lysates for the patients identified above each lane. CTRL, control, NES, nuclear export signal; NLS1, nuclear localization signal 1; NLS2, nuclear localization signal 2; NR, not reported.