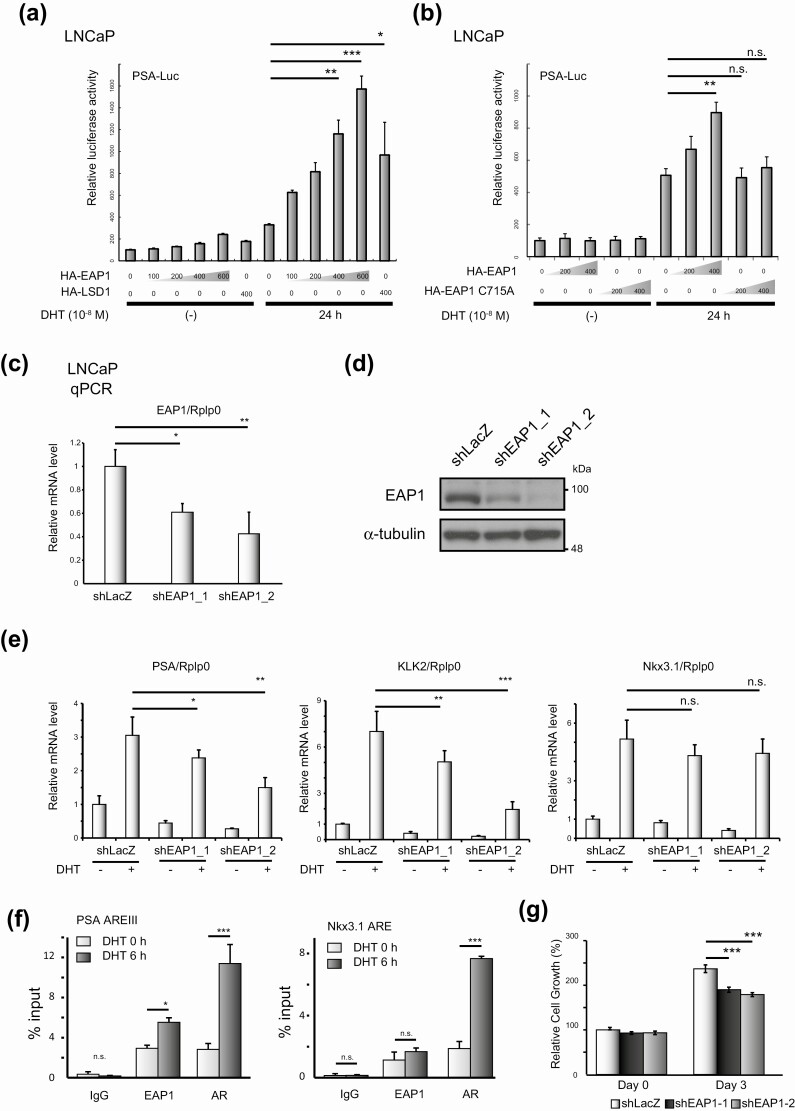
Figure 3.
Enhanced at puberty 1 (EAP1) is an androgen receptor (AR) coactivator. A, LNCaP cells were transiently transfected with the pGL4.1-PSA-Luc reporter (500 ng) and pcDNA3.1-hemagglutinin (HA)-EAP1 expression vectors at the indicated amounts (ng). After 24 hours of incubation, the cells were stimulated with vehicle (ethanol) or 10-nM dihydrotestosterone (DHT) for 24 hours. Data were analyzed by one-way analysis of variance (ANOVA) with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01; ***P less than .001. HA-tagged lysine-specific demethylase 1 (LSD1), as an AR coactivator, was used as the positive control. B, LNCaP cells were transiently transfected with the pGL4.1-PSA-Luc reporter vector (500 ng) and pcDNA3.1 expressing either HA-tagged wild-type EAP1 or HA-tagged EAP1 C715A mutant at the indicated amounts. After 24 hours’ incubation, cells were stimulated with vehicle (ethanol) or 10-nM DHT for 24 hours. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. **P less than .01; n.s., not significant. C, Knockdown of EAP1 by short hairpin RNA (shRNA). Quantitative polymerase chain reaction (qPCR) analysis for EAP1 was performed. LNCaP cells expressing shLacZ were used as a negative control. Gene expression was normalized to that of Rplp0 and presented as the fold change in expression relative to the expression level in cells not treated with DHT. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01. E, Knockdown of EAP1 by shRNA and its effect on the transcriptional activity of AR. Cells were treated with 10-nM DHT for 12 hours, and qPCR analysis of AR target genes was performed. Gene expression was normalized to that of Rplp0 and presented as the fold change in expression relative to the expression level in cells not treated with DHT. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01; ***P less than .001; n.s., not significant. F, Chromatin immunoprecipitation analysis of EAP1 and AR at the indicated gene enhancers. DNA fragments in LNCaP cells were precipitated with anti-EAP1 and anti-AR; rabbit immunoglobulin G (IgG) was used as a negative control for the immunoprecipitation. Precipitated DNA fragments were assessed by qPCR. The error bars indicate SDs. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 3) Error bars indicate SDs. *P less than .05; ***P less than .001; n.s., not significant. G, Cell proliferation assays were performed using the WST-8 cell proliferation assay. The relative absorbance of each cell at the indicated time points is shown. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 5) Error bars indicate SDs. ***P less than .001.