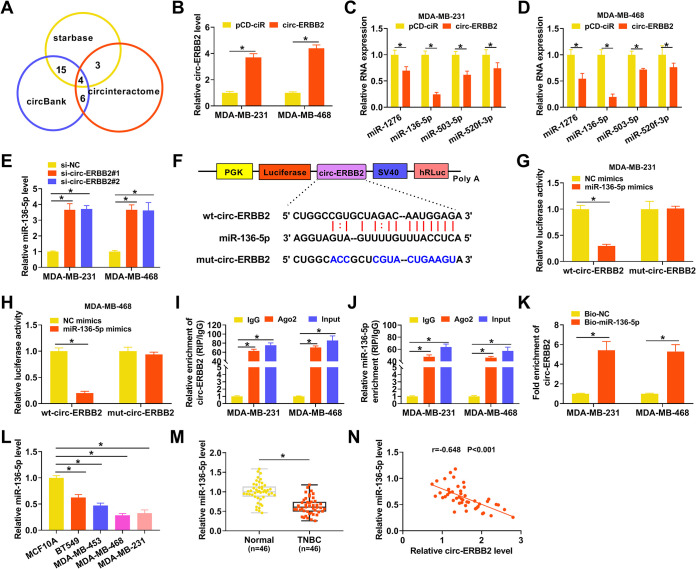
FIG 3.
circ-ERBB2 served as a sponge for miR-136-5p in TNBC cells. (A) Venn diagram showing miRs with a complementary binding sequence to circ-ERBB2 in the circinteractome, circBank, and starbase databases. (B) The relative expression of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells transfected with pCD-ciR or -circ-ERBB2 was measured by qRT-PCR. (C and D) The influence of circ-ERBB2 overexpression on the expression of miR-1276, miR-136-5p, miR-503-5p, and miR-520f-3p in MDA-MB-231 and MDA-MB-468 cells was determined by qRT-PCR. (E) The impact of circ-ERBB2 inhibition on the expression of miR-136-5p in MDA-MB-231 and MDA-MB-468 cells was assessed by qRT-PCR. (F) Schematic illustration of a luciferase reporter containing wt-circ-ERBB2 or mut-circ-ERBB2. SV40, simian virus 40. (G and H) MDA-MB-231 and MDA-MB-468 cells were cotransfected with the luciferase reporter containing wt-circ-ERBB2 or mut-circ-ERBB2 and miR-136-5p mimics or NC mimics, and luciferase activity was analyzed by a dual-luciferase reporter assay. (I and J) The enrichment of circ-ERBB2 and miR-136-5p in immunoprecipitates of the Ago2 and IgG groups was analyzed by qRT-PCR. (K) After an RNA pulldown assay, the abundance of circ-ERBB2 in the Bio-miR-136-5p and Bio-NC groups was analyzed by qRT-PCR. (L and M) The relative expression of miR-136-5p in TNBC cells and tissues was evaluated using qRT-PCR. (N) Pearson’s correlation analysis revealed the correlation between circ-ERBB2 and miR-136-5p in TNBC tissues. PGK, phosphoglycerate kinase; hRLuc, Renilla luciferase. *, P < 0.05.