FIG 1.
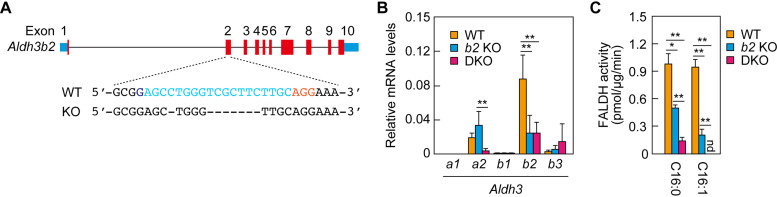
Creation of Aldh3a2 Aldh3b2 double-knockout (DKO) mice and analysis of fatty aldehyde dehydrogenase (FALDH) activity. (A) Aldh3b2 knockout (KO) mice were generated using the CRISPR/Cas9 system. The exon structure (red, coding sequence; blue, untranslated regions) of Aldh3b2 and the nucleotide sequences of wild-type (WT) and Aldh3b2 KO mice around the guide RNA target sequence (light blue) and the protospacer flanking motif (PAM) sequence (magenta) in exon 2 are shown. (B) Total RNA prepared from the epidermis of WT, Aldh3b2 KO, and DKO mice was subjected to SYBR green-based quantitative real-time reverse transcription-PCR (RT-PCR) using specific primers for Aldh3a1, Aldh3a2, Aldh3b1, Aldh3b2, and Aldh3b3. Values are expressed relative to Gapdh and are the means ± standard deviation (SD) of three independent experiments. Statistically significant differences are indicated (**, P < 0.01; Tukey’s test). (C) Total lysates (15 μg) prepared from the epidermis of WT, Aldh3b2 KO, and DKO mice were incubated with 1.5 mM NAD+ and 100 μM hexadecanal (C16:0) or trans-2-hexadecenal (C16:1) at 37°C for 30 min. The quantity of NADH product was determined by measuring the fluorescence of NADH (461.5 nm). Values presented are the means ± SD of three independent experiments. Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01; Tukey’s test). b2 KO, Aldh3b2 KO; nd, not detected.