Abstract
PURPOSE
This study sought to determine the prognostic significance of the WHO-defined glioma molecular subgroups along with additional alterations, including MGMT promoter methylation and mutations in ATRX, CIC, FUBP1, TERT, and TP53, in NRG/RTOG 0424 using long-term follow-up data.
METHODS
Mutations were determined using an Ion Torrent sequencing panel. 1p/19q co-deletion and MGMT promoter methylation were determined by Affymetrix OncoScan and Illumina 450K arrays. Progression-free survival (PFS) and overall survival (OS) were estimated using the Kaplan-Meier method and tested using the log-rank test. Hazard ratios were calculated using the Cox proportional hazard model. Multivariable analyses (MVAs) included patient pretreatment characteristics.
RESULTS
We obtained complete molecular data to categorize 80/129 eligible patients within the WHO subgroups. Of these, 26 (32.5%) were IDHmutant/co-deleted, 28 (35%) were IDHmutant/non-co-deleted, and 26 (32.5%) were IDHwild-type. Upon single-marker MVA, both IDHmutant subgroups were associated with significantly better OS and PFS (P values < .001), compared with the IDHwild-type subgroup. MGMT promoter methylation was obtained on 76 patients, where 58 (76%) were methylated and 18 (24%) were unmethylated. Single-marker MVAs demonstrated that MGMT promoter methylation was statistically significant for OS (P value < .001) and PFS (P value = .003). In a multimarker MVA, one WHO subgroup comparison (IDHmutant/co-deleted v IDHwild-type) was significant for OS (P value = .045), whereas MGMT methylation did not retain significance.
CONCLUSION
This study reports the long-term prognostic effect of the WHO molecular subgroups, MGMT promoter methylation, and other mutations in NRG/RTOG 0424. These results demonstrate that the WHO molecular classification and MGMT both serve as strong prognostic indicators, but that MGMT does not appear to add statistically significant prognostic value to the WHO subgrouping, above and beyond IDH and 1p/19q status.
INTRODUCTION
NRG Oncology/RTOG 0424 was a single-arm, phase II study of radiation therapy (RT) plus temozolomide (TMZ) in patients with high-risk, grade II glioma. This study was designed to provide preliminary data for the combinatorial radiotherapy-TMZ regimen, as a prelude to randomized testing. Long-term results from this study demonstrated that survival for patients treated with RT and TMZ was significantly longer compared with a prespecified historical RT-only control group.1 Molecular analyses for patients on NRG/RTOG 0424 revealed MGMT promoter methylation to be a strong prognostic biomarker, independent of other clinical variables as well as IDH1/2 mutation status.2 It is clear from numerous clinical trials of lower-grade gliomas (LGGs, grade II-III) that the addition of alkylating chemotherapy to RT extends survival for these patients.1,3-5 What remains unclear is whether this is true for all molecular subgroups. We recently reported that only the IDHmutant (IDHmut) WHO subgroups (IDHmutant/co-deleted [IDHmut/co-del] and IDHmutant/non-co-deleted [IDHmut/non-co-del]) received therapeutic benefit from the addition of PCV (procarbazine, lomustine (CCNU), and vincristine) chemotherapy to RT in the NRG/RTOG 9802 trial, but whether the same observation could be extended to TMZ-based regimens remained unclear.6 Therefore, using the long-term clinical data, we sought to determine the prognostic significance of the WHO molecular subgroups in NRG/RTOG 0424. Additionally, we investigated the prognostic significance of MGMT promoter methylation as well as other molecular alterations common in LGGs.
CONTEXT
Key Objective
Clinical trial studies that incorporate comprehensive molecular testing remain limited for lower-grade glioma in the context of temozolomide (TMZ) and radiation. This analysis examined the prognostic performance of the WHO-defined glioma molecular subgroups in NRG Oncology/RTOG 0424, a phase II trial of TMZ-based chemoradiation in high-risk, grade II gliomas.
Knowledge Generated
These results demonstrate the prognostic significance of both the WHO molecular classification and MGMT promoter methylation. Importantly, in this long-term study, MGMT does not appear to add significant value to the WHO subgrouping, above and beyond IDH and 1p/19q status.
Relevance
This is the first phase II study, to our knowledge, to validate the prognostic importance of the WHO molecular classification in patients with grade II glioma receiving TMZ and radiation with long-term follow-up. While IDH and 1p/19q are key biomarkers, comprehensive molecular testing (ie, MGMT methylation, TERT mutation, and EGFR amplification, etc) is necessary for clinical decision making.
METHODS
Patient Cohort
We received formalin-fixed paraffin-embedded (FFPE) tissue in the form of either 1-mm core punches or unstained slides from 105/129 (81%) eligible patients in NRG/RTOG 0424. Representative areas (> 70% tumor) were selected for DNA isolation.
DNA Isolation
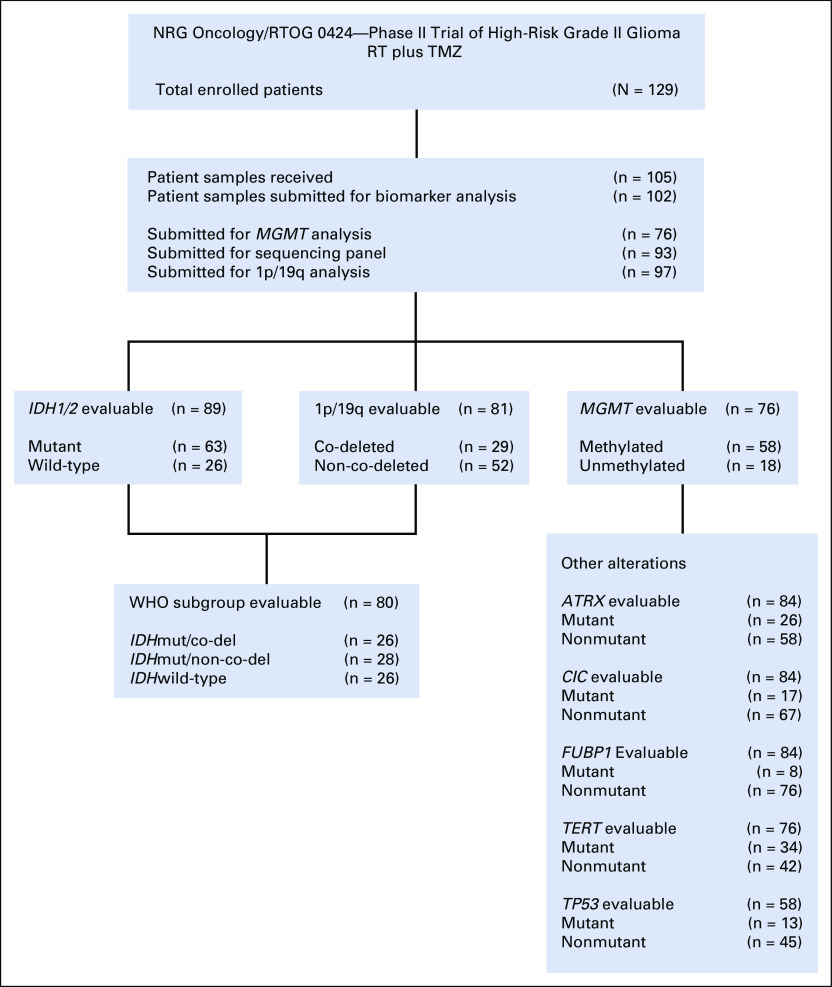
DNA was isolated using an optimized protocol specific for FFPE biospecimens as previously described.6 Figure 1 summarizes the number of patients who had sufficient DNA extracted and were submitted to each of the molecular profiling platforms.
FIG 1.
Molecular biomarker analysis in NRG/RTOG 0424. Sufficient DNA was obtained from 102/105 patients and subsequently submitted to the following platforms in a prioritized manner; (1) MGMT analysis, (2) sequencing panel, (3) 1p/19q analysis. RT, radiation therapy; TMZ, temozolomide.
Mutation and 1p/19q Co-Deletion
A customized Ion AmpliSeq (Thermo Fisher Scientific) sequencing panel was used to assess IDH1, IDH2, ATRX, CIC, and FUBP1 mutation status (Data Supplement). Mutations in the TERT promoter were evaluated using Sanger sequencing. TP53 mutation status was determined using the Affymetrix OncoScan FFPE Assay. 1p/19q co-deletion status was determined using the Affymetrix OncoScan FFPE Assay and the Illumina 450K Methylation Array (Data Supplement).
MGMT Promoter Methylation
MGMT promoter methylation was determined using the Illumina 450K Methylation Array and MGMT status was called using the MGMT-STP27 model.7
Statistical Analysis
Overall survival (OS) and progression-free survival (PFS) by marker status were estimated using the Kaplan-Meier method and tested using the log-rank test. Hazard ratios (HRs) and corresponding 95% CIs were determined using the Cox proportional hazards model. A stepwise model was used for multivariable analyses (MVAs) and the following patient pretreatment characteristics were considered as covariates: sex, histology, age, neurologic function, Zubrod score, tumor crossing the midline, extent of surgery, and tumor size. Subgroup analyses comparing MGMT methylation status within the IDHwild-type (IDHwt) subgroup were conducted but no formal statistical tests were performed because of the small sample size.
RESULTS
Molecular Status
IDH1/2 mutation.
Of the 93 patients who had sufficient DNA for sequencing, 89 (96%) were successfully called for the IDH1/2 mutation, and of these, 63 (71%) patients were IDH1/2 mutant and 26 (29%) were IDH1/2 wild-type. The majority (81%) of the IDH1/2 mutation calls were at the canonical IDH1R132H position (51/63), whereas we also observed five IDH1R132C (8%), two IDH1R132G (3%), three IDH1R132S (5%), and two IDH2R172K (3%) mutations within our patient cohort.
1p/19q co-deletion and WHO classification.
Eighty-one patients had good-quality copy-number data available to call 1p/19q status from either Affymetrix OncoScan arrays (n = 57) or Illumina DNA Methylation 450K arrays (n = 62), whereas 38 patients' data were available from both platforms. Concordance in 1p/19q calls between the Affymetrix and Illumina platforms was 100%. Of these patients, 29 (36%) were 1p/19q co-deleted and 52 (64%) 1p/19q non-co-deleted.
Of the 89 patients with known IDH1/2 mutation status, 1p/19q co-deletion status was available for 73 patients. In this group, 54/73 (74%) were IDH1/2 mutant, of whom 48% (26/54) were 1p/19q co-deleted. The remaining 16 patients with known IDH1/2 mutation status were unable to be called for 1p/19q. Of these, seven patients were IDH1/2 wild-type and thus classified as IDHwt, and nine were IDH1/2 mutant and were designated unclassified.
Therefore, overall, 80 patients had adequate IDH1/2 mutation and 1p19q co-deletion data for classification into the three WHO prognostic subgroups IDHmut/co-del (oligodendroglioma); IDHmut/non-co-del (astrocytoma IDHmut); and IDHwt (astrocytoma IDHwt). Twenty-six (32.5%) patients were classified as IDHmut/co-del, 28 (35%) were IDHmut/non-co-del, and 26 (32.5%) were IDHwt.
MGMT promoter methylation and other alterations.
Seventy-six patients were submitted for Illumina 450K Methylation Array analysis and 58 (76%) were MGMT methylated and 18 (24%) were MGMT unmethylated.
For the 63 patients with IDH1/2 mutations, MGMT status was known for 51 patients, and was methylated in 48 (94%), whereas for the 26 patients who were IDH1/2 wild-type, MGMT status was known for 16 patients and five (45%) patients were methylated.
Sequencing information from ATRX, CIC, and FUBP1 were acquired from the same custom Ion Torrent panel used to obtain IDH1 and IDH2 mutation status. Ninety-three patient samples were sequenced, and good-quality data were obtained on all but nine patients because of low coverage in one or more genes (Data Supplement). Mutations were identified within ATRX in 31% (26/84), CIC in 20% (17/84), and FUBP1 in 10% (8/84) of patients. Specific site mutations within TP53 were available from Affymetrix OncoScan data on 58 patients. Of these, 13 patients (22%) had TP53 mutations (Data Supplement). Other mutations included on the Affymetrix OncoScan array were in BRAF, EGFR, KRAS, NRAS, PIK3CA, and PTEN; however, because of low mutation frequencies, these genes were not used for further correlative analyses. To determine the functional impact of each mutation, a predictive algorithm was used (Mutation Assessor;9 Data Supplement).10
TERT promoter mutations were acquired through Sanger sequencing, and only hotspot mutations at C250T (-146 bp) and C228T (-124 bp) were considered in the final analysis. Of the 76 patients with TERT promoter data, 34 (45%) were mutant and 42 (55%) were nonmutant. 26% (9/34) of those mutated had the C250T-146bp site alteration and 74% (25/34) of patients had a mutation at the C228T-124bp site.
The complete molecular landscape of patients included in these analyses can be found in Figure 2, and mutation diagrams for each gene are demonstrated in the Data Supplement.
FIG 2.

Molecular landscape in NRG/RTOG 0424. A summary of the molecular findings in 98 NRG/RTOG 0424 cases along with select clinical data including age, sex, and histology. The top row shows the classification of patients into the three established molecular subgroups (IDHmutant/co-deleted [IDHmut/co-del], IDHmutant/non-co-deleted [IDHmut/non-co-del], and IDHwild-type [IDHwt]), along with a fourth group, unclassified, because of the lack of available information within these patients. ND, not determined.
Prognostic Correlative Analyses
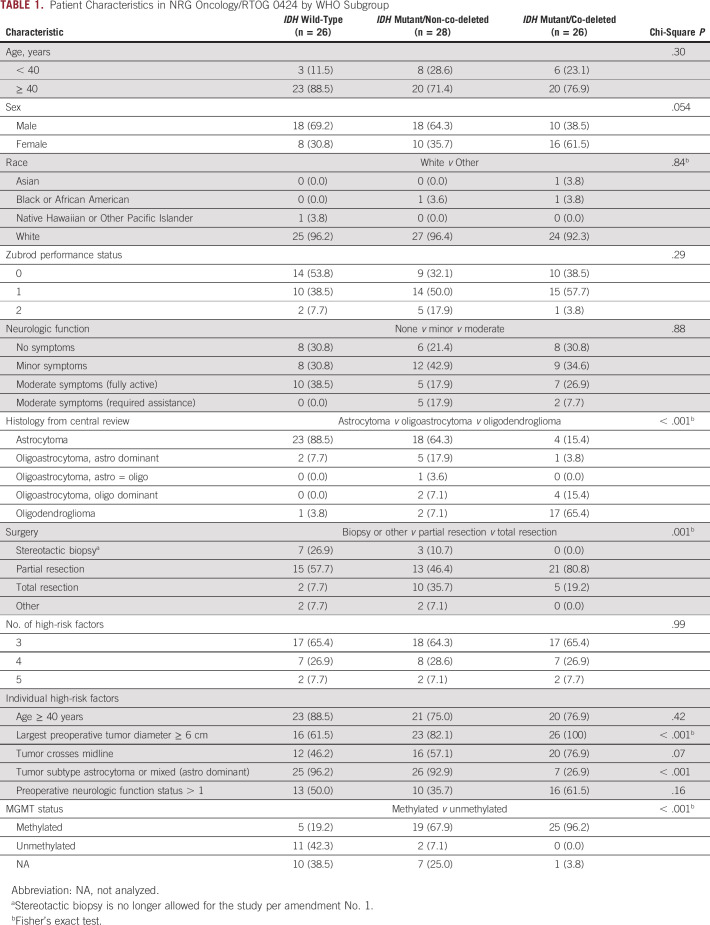
Median follow-up time was 6.9 years (range, 0.3-11.8 years) for all eligible patients in this analysis (n = 98) and 8.8 years (range, 4.5-11.8 years) for all eligible patients alive at the time of analysis (n = 41). Pretreatment characteristics were not significantly different between patients included in this analysis versus those not included, except for surgery (P = .01; Data Supplement). Patient characteristics by WHO molecular subgroup are shown in Table 1. Extent of surgery and tumor size were highly imbalanced and thus not considered as covariates for Cox proportional hazards models.
TABLE 1.
Patient Characteristics in NRG Oncology/RTOG 0424 by WHO Subgroup
Univariable analyses.
When comparing the three WHO molecular subgroups, we found that two of the three comparisons (IDHmut/co-del v IDHwt and IDHmut/non-co-del v IDHwt) were statistically significant for OS (Table 2; Fig 3A), but the comparison between IDHmut/co-del versus IDHmut/non-co-del was not significantly different. The median survival times (MSTs) were 9.4 years (95% CI, 8.2 to not reached [NR]; IDHmut/co-del), 8.8 years (95% CI, 5.9 to NR; IDHmut/non-co-del), and 2.3 years (95% CI, 1.4 to 3.4; IDHwt). Patients with MGMT methylated tumors had significantly better OS (HR, 0.31; 95% CI, 0.17 to 0.58; P < .001), with MSTs of 9.0 years (95% CI, 8.1 to NR), compared with 3.0 years (95% CI, 2.2 to 5.9) for MGMT unmethylated tumors (Fig 4A). A subgroup analysis looking at MGMT methylation status within the IDHwt subgroup visually showed no major difference in OS, where the MST for IDHwt/MGMT methylated patients was 3.8 years (95% CI, 0.7 to NR) and 2.6 years (95% CI, 1.4 to 4.4) for IDHwt/MGMT unmethylated patients (Data Supplement). As individual biomarkers, IDH1/2 mutation and 1p19q co-deletion were each significantly correlated with OS (Data Supplement).
TABLE 2.
Univariable and Single-Marker Multivariable Cox Proportional Hazards Models for OS and PFS With WHO Subgroup
FIG 3.
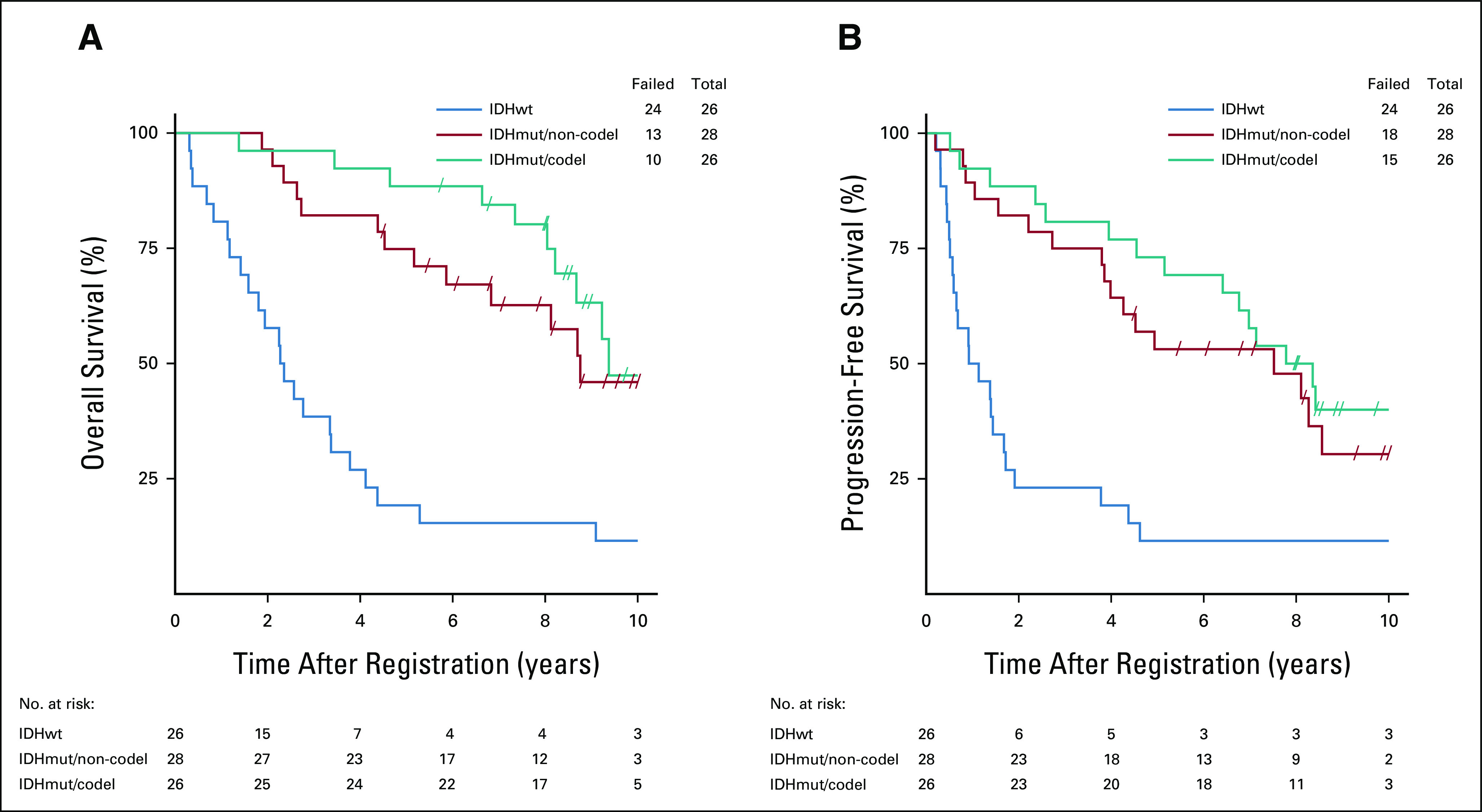
Survival by WHO molecular subgroup. Kaplan-Meier survival plots demonstrate the three WHO-defined molecular subgroups (IDHmutant/co-deleted [IDHmut/co-del], IDHmutant/non-co-deleted [IDHmut/non-co-del], and IDHwild-type [IDHwt]), and stratified patients for both (A) OS and (B) PFS. OS, overall survival; PFS, progression-free survival.
FIG 4.
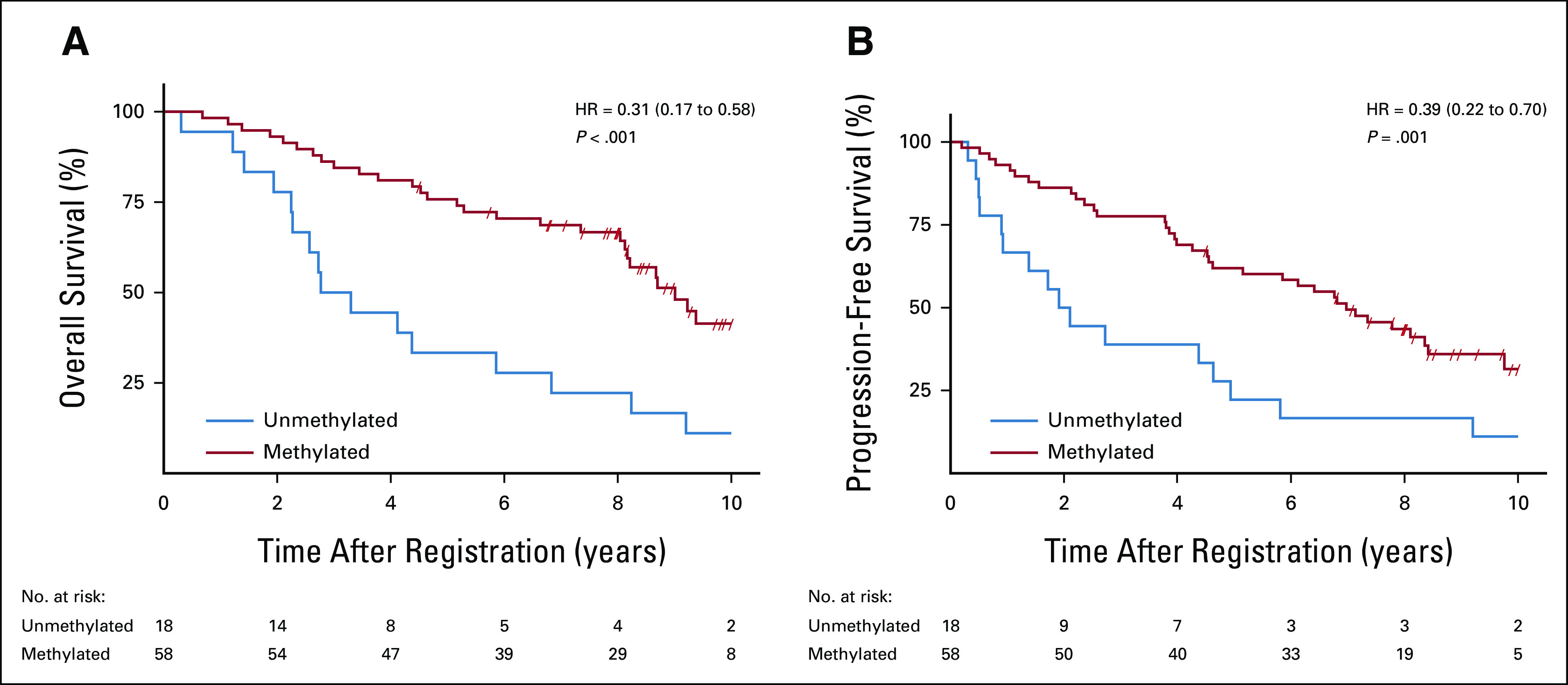
Survival by MGMT promoter methylation. Kaplan-Meier survival plots show that patients with MGMT methylated tumors experienced significantly longer (A) OS and (B) PFS rates compared to patients with MGMT unmethylated tumors. OS, overall survival; PFS, progression-free survival.
For PFS, each IDHmut subgroup was significantly associated with better outcome compared with the IDHwt group (Table 2; Fig 3B). The median PFS times were 8.1 years (95% CI, 5.2 to NR; IDHmut/co-del), 7.5 years (95% CI, 3.9 to 11.8; IDHmut/non-co-del), and 1.0 year (95% CI, 0.6 to 1.7; IDHwt). MGMT promoter methylation was also significantly associated with better PFS (HR, 0.39; 95% CI, 0.22 to 0.70; P = .001; Fig 4B). Median PFS times were 7.0 years (95% CI, 4.5 to 8.4) for patients with MGMT methylated tumors and 2.0 years (95% CI, 0.9 to 4.6) for those with MGMT unmethylated tumors. Subgroup analysis looking at MGMT methylation status within the IDHwt subgroup showed visual trends for methylated patients having better PFS, with the median PFS time for IDHwt/MGMT methylated patients at 3.8 years (95% CI, 0.7 to NR) and for IDHwt/MGMT unmethylated patients at 1.4 years (95% CI, 0.4 to 4.4) (Data Supplement). For PFS, IDH1/2 mutation and 1p/19q co-deletion were each significantly associated with better PFS as individual biomarkers (Data Supplement).
None of the other mutations evaluated (ATRX, CIC, FUBP1, TERT promoter, and TP53) were found to be significantly associated with OS or PFS (Data Supplement).
Single-marker MVAs.
For single-marker MVA, the following factors were included: sex, histology, age, neurologic function, Zubrod score, and tumor crossing the midline. Statistical significance for the two WHO subgroup comparisons was maintained in MVAs for OS (IDHmut/co-del v IDHwt; HR, 0.18 [95% CI, 0.08 to 0.38]; P < .001 and IDHmut/non-co-del v IDHwt; HR, 0.25 [95% CI, 0.12 to 0.50]; P < .001; Table 2). Upon MVA, MGMT promoter methylation was found to be associated with significantly better OS (HR, 0.33; 95% CI, 0.18 to 0.61; P < .001; Data Supplement). As individual biomarkers, significance was maintained in MVAs for IDH1/2 mutation and 1p/19q co-deletion (Data Supplement).
For PFS, the WHO subgroup analyses remained significant upon MVA (IDHmut/co-del v IDHwt; HR, 0.25 [95% CI, 0.13 to 0.49]; P < .001 and IDHmut/non-co-del v IDHwt; HR, 0.31; 95% CI, 0.17 to 0.59; P < .001; Table 2). MGMT status also remained associated with significantly better PFS (HR, 0.40; 95% CI, 0.22 to 0.73; P = .003; Data Supplement). As individual biomarkers, IDH1/2 mutation and 1p/19q co-deletion retained significance (Data Supplement).
Multimarker MVAs.
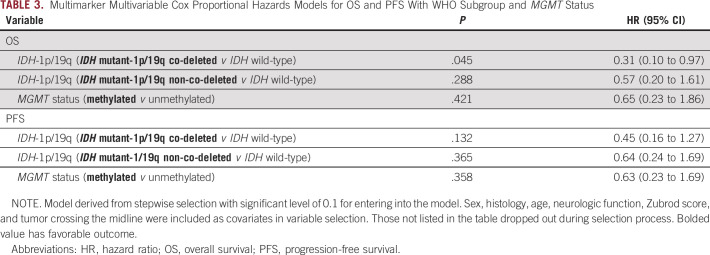
For multimarker MVA, the following factors and markers were included: WHO molecular subgroup, MGMT methylation, sex, histology, age, neurologic function, Zubrod score, and tumor crossing the midline. This resulted in the following comparative groups: IDHmut/co-del/MGMT methylated (25 patients), IDHmut/co-del/MGMT unmethylated (0 patients), IDHmut/non-co-del/MGMT methylated (19 patients), IDHmut/non-co-del/MGMT unmethylated (2 patients), IDHwt/MGMT methylated (five patients), and IDHwt/MGMT unmethylated (11 patients). In this multimarker MVA, one WHO subgroup comparison (IDHmut/co-del v IDHwt) was associated with significantly better OS (HR, 0.31; 95% CI, 0.10 to 0.97; P = .045) and trended with PFS (HR, 0.45; 95% CI, 0.16 to 1.27; P = .13), whereas the effect of MGMT methylation was not significant for either OS or PFS (Table 3).
TABLE 3.
Multimarker Multivariable Cox Proportional Hazards Models for OS and PFS With WHO Subgroup and MGMT Status
DISCUSSION
In this study, we demonstrate that patients with high-risk, IDH1/2 mutant grade II glioma, regardless of 1p/19q co-deletion status, experienced significantly longer survival when treated with RT plus TMZ relative to IDHwt patients. These results are similar to those reported for NRG/RTOG 98026; however, in NRG/RTOG 0424, a significant survival benefit in patients with IDHmut/co-del tumors over those in the IDHmut/non-co-del subgroup was not observed, although this may be because of our small sample size or the imbalance of clinical features between subgroups. Additionally, recent results from the CATNON trial showed no overall benefit from the addition of concurrent or adjuvant TMZ to RT in IDHwt anaplastic glioma.9 Our study supports the notion that survival outcomes for grade II IDHwt patients remain dismal (MST = 2.3 years) and comparable to grade III anaplastic astrocytomas.10 While these results confirm the prognostic findings of numerous other comprehensive LGG studies,3,11-14 our study used tissue that was prospectively collected from high-risk, grade II patients treated on a phase II trial. Overall, these findings suggest that IDHwt patients should be separated from IDHmut patients in future grade II glioma trials, and novel treatment strategies are warranted.
As previously shown in our short-term report,2 MGMT promoter methylation remains a highly significant prognostic biomarker for OS and PFS in both univariable and single-marker MVAs including clinical factors using the long-term clinical follow-up data. However, the significance of MGMT was not retained in the long-term multimarker MVAs that included WHO molecular subgroups, which may be because of the small sample size or the dependency of MGMT promoter methylation on IDH1/2 mutation. Median follow-up for patients in this analysis was 6.9 years, whereas for the short-term report, median follow-up was 4.1 years.15 Although our previous report suggested that MGMT may add value to the IDHwt subgroup,2 analyses using the long-term follow-up data visually showed no difference in survival outcomes between methylated and unmethylated patients (Data Supplement). These findings need to be interpreted with caution and require further validation in a larger cohort. Unfortunately, the IDHmut molecular subgroups were too small to determine whether MGMT promoter methylation added prognostic value. Other LGG studies have also shown IDH1/2 mutation to be a stronger prognostic marker than MGMT promoter methylation6,16; however, as evidenced by the cIMPACT-NOW updates,17-19 comprehensive molecular testing (ie, IDH1/2, 1p/19q, MGMT methylation, TERT, EGFR, and chr7/9/10) is necessary for accurate classification and assigning the most appropriate therapeutic regimen to patients with LGG.
When we assessed the prognostic significance of additional gene mutations, none were found to be statistically significant (Data Supplement). We did, however, observe trends for both CIC and FUBP1 mutations being associated with better OS and PFS, although these alterations commonly coincide with 1p/19q co-deletions (Data Supplement). Similar to other studies, in this cohort, we predominantly observed ATRX and TP53 mutations in the IDHmut/non-co-del subgroup,13,20,21 CIC and FUBP1 mutations in IDHmut/co-del patients,13,21 and TERT promoter mutations within both IDHmut/co-del and IDHwt patients.13,20,22 Although we did perform subset analyses evaluating the prognostic significance of each mutation within the WHO molecular subgroups, none were significant (P > .2). Again, subset sample sizes were too small for meaningful interpretation (Data Supplement).
In NRG/RTOG 0424, the addition of TMZ to RT was shown to significantly improve survival outcomes for patients compared with the RT-only historical control group.1,15 To better understand which of the WHO molecular subgroups benefit from the addition of TMZ to RT, we performed a post hoc cross-trial comparison of NRG/RTOG 0424 and the RT-alone arm of NRG/RTOG 9802.6 This cross-trial comparison, however, has limitations because of small subset sample sizes and differences in patient pretreatment characteristics between the two trials. NRG/RTOG 0424 consisted of patients who were older, had slightly more neurologic symptoms, were of astrocytoma histology, and had a different distribution of surgery. For OS, no difference was observed within the IDHwt subgroup, but IDHmut/non-co-del patients experienced longer median OS times with the addition of TMZ to RT (8.8 years) compared with RT-alone (4.3 years). Interestingly, IDHmut/co-del patients treated with RT-alone experienced longer median OS times (13.9 years) compared to those treated with RT plus TMZ (9.4 years), which may be because of differences in the clinical characteristics mentioned above. For PFS, all subgroups experienced longer MSTs when treated with RT plus TMZ compared to those treated with RT-alone (Data Supplement).6
Over the past decade, there has been an ongoing debate regarding the usage of PCV versus TMZ in the setting of LGG. From previous studies, patients treated on NRG/RTOG 9802 (RT plus PCV) experienced longer median OS (13.3 years) and PFS (10.4 years) times compared with those treated on NRG/RTOG 0424 (OS, 8.2 years; PFS, 4.5 years; Data Supplement).1,5 Again, these findings may be because of differences within patient populations as these trials defined high risk differently, and in general, NRG/RTOG 0424 contained more patients with high-risk features.
When comparing the WHO molecular subgroups between NRG/RTOG 9802 (RT plus PCV arm) and NRG/RTOG 0424, we observed similar median OS times within the IDHwt subgroup (2.1 v 2.3 years); however, extended OS survival times were observed for each of the IDHmut subgroups when treated with RT plus PCV versus RT plus TMZ (IDHmut/co-del; [NR v 9.4 years] and IDHmut/non-co-del [11.4 v 8.8 years]). We observed a similar effect within subgroups when looking at median PFS times (Data Supplement). The added benefit of PCV, specifically within the IDHmut/co-del subgroup, has been observed in other trials, suggesting this effect may be inherent to oligodendrogliomas.3,4,6,12
The small sample size of the NRG/RTOG 0424 study is a clear limitation to obtaining appropriate power for certain molecular subsets and MVAs. A significantly lower proportion of patients with biopsy only had specimens available for use as compared to those without available specimens (10.2% v 32.3%; P = .01), which may compromise the generalizability of these results to the study as a whole. Additionally, results may have been affected by the imbalance of surgery and tumor size between subgroups.
Additional underlying molecular mechanisms (ie, microvascular proliferation, and necrosis, etc) may be contributing to a more aggressive phenotype in NRG/RTOG 0424, and as recommended by cIMPACT, further molecular testing can aid in the clinical management of LGGs.17-19 For example, combined chemoradiotherapy is recommended for IDHwt anaplastic astrocytoma and diffuse astrocytoma if certain molecular features are present (ie. EGFR amplification, gain of chromosome 7, loss of 10, or TERT promoter mutation).17 Ongoing trials (CODEL and CATNON) will help clinicians develop more personalized treatment plans for LGG, thereby improving survival outcomes for these patients. Notably, novel treatment approaches are desperately needed for high-risk LGGs, specifically for the IDHwt subgroup.
ACKNOWLEDGMENT
The authors thank the USC Epigenome Center, NRG Oncology Biorepository, and The Ohio State Comprehensive Cancer Center Solid Tumor and Biostatistics Cores (supported in part by grant P30 CA016058 from the NCI). In addition, the authors thank Ziyan Liu, MS, Johnson & Johnson, for assistance in DNA isolation and database organization. A.C. and E.H.B. also acknowledge the Cancer Therapy Evaluation Program of the National Cancer Institute (NRG-BN-TS006, to A.C. and E.H.B.).
Stephanie L. Pugh
Research Funding: Pfizer, Millennium
Glenn J. Lesser
Honoraria: SDP Oncology
Consulting or Advisory Role: Cancer Expert Now, Agios, Incysus
Research Funding: Novocure, Oblato, Denovo Biopharma, Global Coalition for Adaptive Research
Other Relationship: NCI, ASCO
David R. Macdonald
Research Funding: Celgene, Servier
Erica H. Bell
Patents, Royalties, Other Intellectual Property: US20180002762A1
Joseph P. McElroy
Employment: Pfizer
Cynthia D. Timmers
Employment: Incyte
Stock and Other Ownership Interests: Array BioPharma, Seattle Genetics, Exact Sciences, Incyte, Arbutus Biopharma, PDS Biotechnology
Consulting or Advisory Role: Ventana Medical Systems
C. Leland Rogers
Employment: Barrow Neurological Institute, GammaWest Cancer Services
Stock and Other Ownership Interests: GT Technologies
Maria Werner-Wasik
Stock and Other Ownership Interests: Illumina
Honoraria: AstraZeneca
Patents, Royalties, Other Intellectual Property: Signal transduction inhibitor in lymphoma
Hsiang-Hsuan Michael Yu
Honoraria: UpToDate, Elsevier, Sermo, Guidepoint Global
Consulting or Advisory Role: Novocure
Speakers' Bureau: BrainLAB
Research Funding: Bristol Myers Squibb/Sanofi, Merck
Travel, Accommodations, Expenses: BrainLAB
David P. D'Souza
Consulting or Advisory Role: AbbVie
Nadia N. Laack
Research Funding: Bristol Myers Squibb
Minesh P. Mehta
Leadership: Oncoceutics
Stock and Other Ownership Interests: Chimerix
Consulting or Advisory Role: Tocagen, Karyopharm Therapeutics, Mevion Medical Systems, ZappRx, Sapience Therapeutics
Patents, Royalties, Other Intellectual Property: WARF patent 14/934,27, Topical Vasoconstritor Preparations and Methods for Protecting Cells During Cancer Chemotherapy and Radiotherapy
Uncompensated Relationships: Xcision Medical Systems, ViewRay
Arnab Chakravarti
Research Funding: Varian Medical Systems
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented at the 2016 Annual Society for Neuro-Oncology (SNO) Meeting, Scottsdale, AZ; and the 2020 Annual American Society of Clinical Oncology (ASCO) Meeting (virtual).
SUPPORT
U10CA180868 (NRG Oncology Operations), U10CA180822 (NRG Oncology SDMC), and U24CA196067 (NRG Specimen Bank), from the National Cancer Institute (NCI) and Merck. This project is funded, in part, under a grant with the Pennsylvania Department of Health (CURE grant). The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. Also, R01CA108633, R01CA169368, RC2CA148190, U10CA180850-01 (NCI), Brain Tumor Funders Collaborative Grant, and the Ohio State University CCC (all to A.C.).
AUTHOR CONTRIBUTIONS
Conception and design: Jessica L. Fleming, Glenn J. Lesser, David R. Macdonald, Erica H. Bell, Kenneth D. Aldape, Arnab Chakravarti
Administrative support: Erica H. Bell, Minesh P. Mehta
Provision of study materials or patients: Glenn J. Lesser, David R. Macdonald, C. Leland Rogers, Thomas J. Doyle, Maria Werner-Wasik, David P. D'Souza, Nadia N. Laack, Penny K. Sneed, Minesh P. Mehta
Collection and assembly of data: Jessica L. Fleming, Stephanie L. Pugh, Barbara J. Fisher, Glenn J. Lesser, David R. Macdonald, Erica H. Bell, Cynthia D. Timmers, C. Leland Rogers, Thomas J. Doyle, Maria Werner-Wasik, Jean-Paul Bahary, David P. D'Souza, Penny K. Sneed, Minhee Won, Minesh P. Mehta, Arnab Chakravarti
Data analysis and interpretation: Jessica L. Fleming, Stephanie L. Pugh, David R. Macdonald, Erica H. Bell, Joseph P. McElroy, Aline P. Becker, Cynthia D. Timmers, Kenneth D. Aldape, C. Leland Rogers, Jean-Paul Bahary, Hsiang-Hsuan Michael Yu, David P. D'Souza, Nadia N. Laack, Young Kwok, Minhee Won, Minesh P. Mehta, Arnab Chakravarti
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stephanie L. Pugh
Research Funding: Pfizer, Millennium
Glenn J. Lesser
Honoraria: SDP Oncology
Consulting or Advisory Role: Cancer Expert Now, Agios, Incysus
Research Funding: Novocure, Oblato, Denovo Biopharma, Global Coalition for Adaptive Research
Other Relationship: NCI, ASCO
David R. Macdonald
Research Funding: Celgene, Servier
Erica H. Bell
Patents, Royalties, Other Intellectual Property: US20180002762A1
Joseph P. McElroy
Employment: Pfizer
Cynthia D. Timmers
Employment: Incyte
Stock and Other Ownership Interests: Array BioPharma, Seattle Genetics, Exact Sciences, Incyte, Arbutus Biopharma, PDS Biotechnology
Consulting or Advisory Role: Ventana Medical Systems
C. Leland Rogers
Employment: Barrow Neurological Institute, GammaWest Cancer Services
Stock and Other Ownership Interests: GT Technologies
Maria Werner-Wasik
Stock and Other Ownership Interests: Illumina
Honoraria: AstraZeneca
Patents, Royalties, Other Intellectual Property: Signal transduction inhibitor in lymphoma
Hsiang-Hsuan Michael Yu
Honoraria: UpToDate, Elsevier, Sermo, Guidepoint Global
Consulting or Advisory Role: Novocure
Speakers' Bureau: BrainLAB
Research Funding: Bristol Myers Squibb/Sanofi, Merck
Travel, Accommodations, Expenses: BrainLAB
David P. D'Souza
Consulting or Advisory Role: AbbVie
Nadia N. Laack
Research Funding: Bristol Myers Squibb
Minesh P. Mehta
Leadership: Oncoceutics
Stock and Other Ownership Interests: Chimerix
Consulting or Advisory Role: Tocagen, Karyopharm Therapeutics, Mevion Medical Systems, ZappRx, Sapience Therapeutics
Patents, Royalties, Other Intellectual Property: WARF patent 14/934,27, Topical Vasoconstritor Preparations and Methods for Protecting Cells During Cancer Chemotherapy and Radiotherapy
Uncompensated Relationships: Xcision Medical Systems, ViewRay
Arnab Chakravarti
Research Funding: Varian Medical Systems
No other potential conflicts of interest were reported.
REFERENCES
- 1.Fisher BJ, Pugh SL, Macdonald DR, et al. Phase 2 study of a temozolomide-based chemoradiation therapy regimen for high-risk, low-grade gliomas: Long-term results of Radiation Therapy Oncology Group 0424 Int J Radiat Oncol Biol Phys 107720–7252020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell EH, Zhang P, Fisher BJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with radiotherapy and temozolomide: An analysis from the NRG Oncology/RTOG 0424 trial JAMA Oncol 41405–14092018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: Long-term follow-up of EORTC Brain Tumor Group Study 26951 J Clin Oncol 31344–3502013 [DOI] [PubMed] [Google Scholar]
- 4.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: Long-term results of RTOG 9402 J Clin Oncol 31337–3432013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckner JC, Shaw EG, Pugh SL, et al. Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma N Engl J Med 3741344–13552016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell EH, Zhang P, Shaw EG, et al. Comprehensive genomic analysis in NRG Oncology/RTOG 9802: A phase III trial of radiation versus radiation plus procarbazine, lomustine (CCNU), and vincristine in high-risk low-grade glioma J Clin Oncol 383407–34172020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status Acta Neuropathol 124547–5602012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Bent MJ, Tesileanu CMS, Wick W, et al. Adjuvant and concurrent temozolomide for 1p/19q non-co-deleted anaplastic glioma (CATNON; EORTC study 26053-22054): Second interim analysis of a randomised, open-label, phase 3 study Lancet Oncol 22813–8232021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang S, Zhang P, Cairncross JG, et al. Phase III randomized study of radiation and temozolomide versus radiation and nitrosourea therapy for anaplastic astrocytoma: Results of NRG Oncology RTOG 9813 Neuro Oncol 19252–2582017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumert BG, Hegi ME, van den Bent MJ, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): A randomised, open-label, phase 3 intergroup study Lancet Oncol 171521–15322016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wick W, Roth P, Hartmann C, et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide Neuro Oncol 181529–15372016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Netwok. Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas N Engl J Med 3722481–24982015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary Acta Neuropathol 131803–8202016 [DOI] [PubMed] [Google Scholar]
- 15.Fisher BJ, Hu C, Macdonald DR, et al. Phase 2 study of temozolomide-based chemoradiation therapy for high-risk low-grade gliomas: Preliminary results of Radiation Therapy Oncology Group 0424 Int J Radiat Oncol Biol Phys 91497–5042015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide J Clin Oncol 275874–58802009 [DOI] [PubMed] [Google Scholar]
- 17.Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV” Acta Neuropathol 136805–8102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas Acta Neuropathol 139603–6082020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading Brain Pathol 30844–8562020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Zhang X, Yan X, et al. Significance of TERT and ATRX mutations in glioma Oncol Lett 1795–1022019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas Nat Genet 47458–4682015 [DOI] [PubMed] [Google Scholar]
- 22.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors N Engl J Med 3722499–25082015 [DOI] [PMC free article] [PubMed] [Google Scholar]