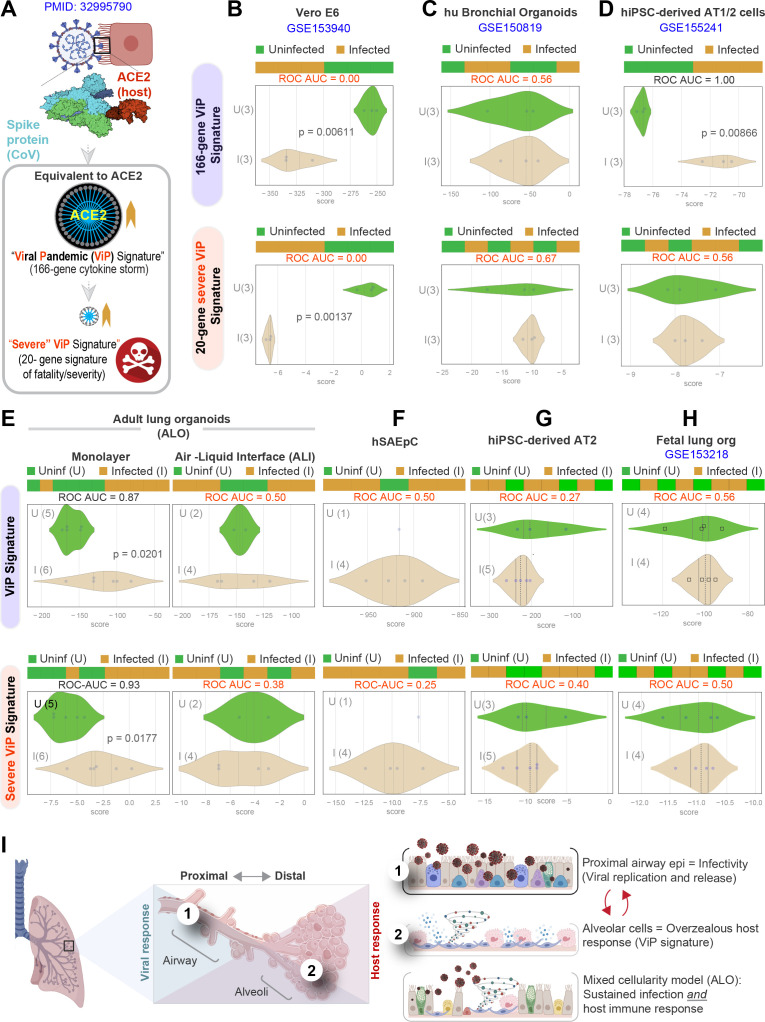
Figure 6. Both proximal and distal airway components are required to model the overzealous host response in COVID-19.
(A) Schematic summarizing the immune signatures identified based on ACE2-equivalent gene induction observed invariably in any respiratory viral pandemic. The 166-gene ViP signature captures the cytokine storm in COVID-19, whereas the 20-gene subset severe ViP signature is indicative of disease severity/fatality. (B–D) Publicly available RNA seq datasets from commonly used lung models, Vero E6 (B), human bronchial organoids (C), and hPSC-derived AT1/2 cell-predominant lung organoids are classified using the 166-gene ViP signature (top row) and 20-gene severity signature (bottom row). (E–G) RNA seq datasets generated in this work using either human small airway epithelial cells (E), adult lung organoids as submerged or air-liquid interphase (ALI) models (left and right, respectively, in F) and hiPSC-derived AT2 cells (G) were analyzed and visualized as in (B–D). (H) Publicly available RNA seq datasets from fetal lung organoid monolayers (Lamers et al., 2021) infected or not with SARS-CoV-2 were analyzed as in (B–D) for the ability of ViP signatures to classify infected (I) from uninfected (U) samples. Receiver operating characteristics area under the curve (ROC AUC) in all figure panels indicate the performance of a classification model using the ViP signatures. (I) Summary of findings in this work, its relationship to the observed clinical phases in COVID-19, and key aspects of modeling the same. Table 6 lists details regarding the patient cohorts/tissue or cell types represented in each transcriptomic dataset.