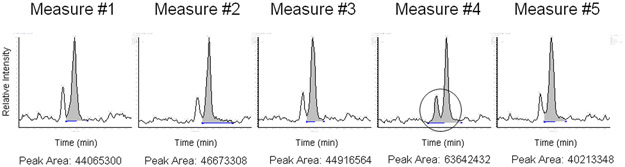
Figure 5.
An example of software analysis variations in MS peak integration when the same peptide (mitochondrial Stress-70 protein) was repeatedly measured. Automatic peak integration and area calculation were performed using the PepQuan module in Bioworks (Rev. 3.3.1 SP1). The extracted ion chromatograms of the peptide were generated from 5 LC-MS/MS analyses using a linear ion trap mass spectrometer LTQ-XL for the same trypsin-digested proteome sample from mouse lymphoma cells (L5178Y/Tk+/−3.7.2C). The circle indicates an additional peak that was included in the calculation of the integrated peak area value. Since these were technical replicates, these results demonstrate another variance that is caused by the post-analytical data processing even though Good Laboratory Practice has been followed and the reproducibility was kept at an optimal level during analytical experiments.