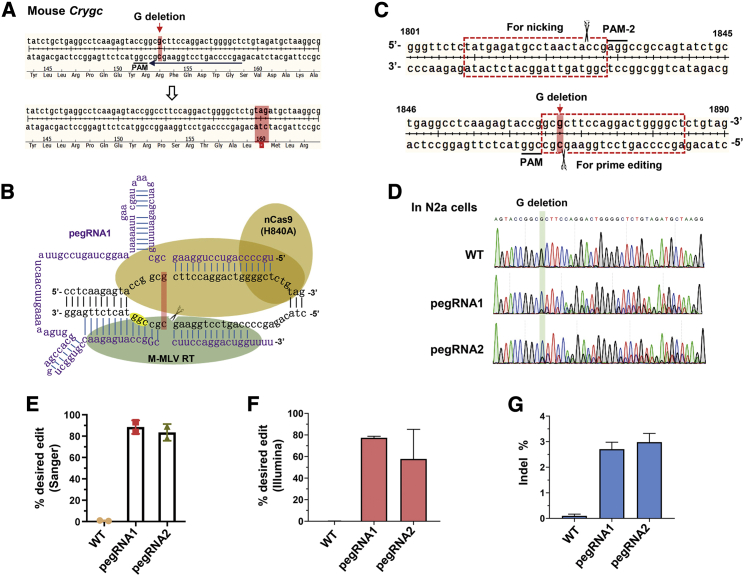
Figure 1.
Highly efficient installation of a G-deletion (G-del) mutation into Crygc with a prime editing system in N2a cells
(A) Schematic diagram showing the target site in mouse Crygc locus. The PAM sequence is underlined in black, and the spacer sequence of pegRNA is underlined in blue. The cataract disorder-associated G nucleotide, the deletion of which results in a premature stop codon (highlighted in the lower panel), is highlighted by a red arrow in the upper panel. (B) The expected structure of PE2 and pegRNA1 used for inducing G-deletion in Crygc. (C) Schematic diagram for the pegRNA spacer (for prime editing) and sgRNA spacer (for nicking) design in the PE3 system for mouse Crygc editing. (D) Editing frequency of PE3 (pegRNA1 and pegRNA2)-induced G-deletion in N2a cells analyzed from Sanger sequencing results. (E) Analysis of editing efficiency from Sanger sequencing results with EditR. Two replicates were independently performed and analyzed. (F) Targeted deep-sequencing analysis of amplicons from two replicates of N2a cells transfected with PE2 and pegRNAs. The proportions of G-deletion reads were presented. (G) The proportions of sequencing reads with indels were presented.