Figure 3.
Applications of MultiEditR to RNA and DNA base editing
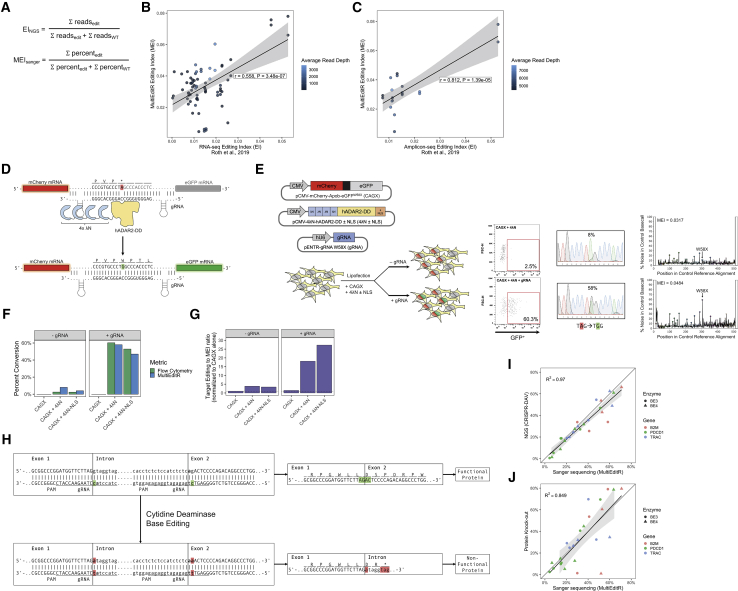
(A) Formula for MultiEditR editing index (MEISanger) and Alu editing index from Roth et al.19 calculated from NGS data (EINGS). (B) Correlation between MEI and RNA-seq EI shaded by average read depth of RNA-seq. (C) Correlation between MEI and Amplicon-seq EI shaded by average read depth of Amplicon-seq. (D) Diagram of 4λN-hADAR2-DD RNA editing system. (E) EGFP reactivation experiment using the 4λN-DD system. Plasmids encoding gRNA, 4λΝ-hADAR2-DD, and mCherry-Apob-EGFPW58X (CAGX) were transfected into HEK293T cells. (F) Reactivation of EGFP measured via flow cytometry and MultiEditR. (G) Ratio of target X58W editing to non-desired events, measured by the ratio of target editing to the MEI. Results are normalized to the CAGX alone negative control. Flow cytometry plots, MultiEditR plots, and analysis parameters are available in Figure S7. (H and I) DNA base editing quantification of rAPOBEC1-BE3 and rAPOBEC1-BE4 in human T cells by MultiEditR compared to Amplicon-seq NGS analyzed by CRISPR-DAV. (H and J) Comparison of MultiEditR to protein KO efficiency as measured by FACS in human T cells; data were analyzed from Webber et al.21