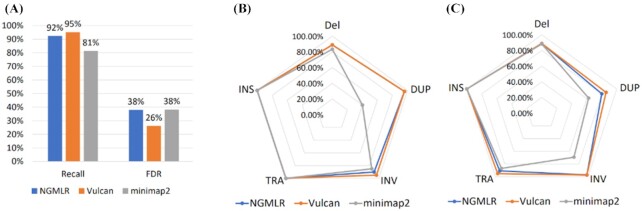
Figure 5:
Benchmarking SV calls on simulated structural variants (INS: insertions; Del: deletions; TRA: translocations; DUP: duplications; INV: inversions) with ONT reads simulated from Saccharomyces cerevisiae. A: Recall and false discovery rate (FDR) of Sniffles’ SV calling on simulated Nanopore reads with three different mappers. SV calls on Vulcan mappings offer the highest recall (95%) and lowest FDR (26%). B: Recall of different SV types from minimap2, NGMLR, and Vulcan mappings with Sniffles’ SV calling on simulated Nanopore reads. C: Precision of different SV types from NGMLR, Vulcan, and minimap2’s mappings with Sniffles’ SV calling on simulated Nanopore reads. NGMLR has similar performance across all SV types, while minimap2 has a lower precision on INVs and DUPs.