Abstract
COVID-19 is a pneumonia-like disease with highly transmittable and pathogenic properties caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which infects both animals and humans. Although many efforts are currently underway to test possible therapies, there is no specific FDA approved drug against SARS-CoV-2 yet. miRNA-directed gene regulation controls the majority of biological processes. In addition, the development and progression of several human diseases are associated with dysregulation of miRNAs. In this regard, it has been shown that changes in miRNAs are linked to severity of COVID-19 especially in patients with respiratory diseases, diabetes, heart failure or kidney problems. Therefore, targeting these small noncoding-RNAs could potentially alleviate complications from COVID-19. Here, we will review the roles and importance of host and RNA virus encoded miRNAs in COVID-19 pathogenicity and immune response. Then, we focus on potential miRNA therapeutics in the patients who are at increased risk for severe disease.
Keywords: SARS-CoV-2, Coronavirus, miRNAs, Anti-miRNA therapy, miRNA therapy
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease (COVID-19), started as a pandemic outbreak on December 2019 in Wuhan, China and rapidly spread across the world. Most patients with COVID-19 tend to have mild symptoms such as low-grade fever, dry cough, and tiredness. However, patients with respiratory failure, type 2 diabetes, heart failure and other cardiovascular diseases or chronic kidney disease (CKD) are at increased risk for severe symptoms [1], [2], [3], [4]. Due to the novelty of COVID-19, there are no specific treatments approved by U.S. Food and Drug Administration (FDA) for this virus; and therefore, there is an urgent need for finding effective and safe therapeutic agents.
MicroRNAs (miRNAs) are a class of endogenously initiated noncoding RNAs (ncRNAs), which are highly conserved between different eukaryotic species and are responsible for gene expression regulation by binding to complementary target mRNAs [5]. miRNAs have autocrine functions but may also be involved in endocrine and paracrine manners through transfer in exosomes [6]. These small RNA molecules can mediate responses to infections especially in anti-viral defense. In addition, miRNAs bind to the 5′- and 3′-untranslated regions (UTRs) of target mRNAs or the amino acid coding sequence (CDS) of the viral genome and regulate their pathogenesis [7]. Notably, there are some miRNAs encoded by RNA viruses that may evolve in regulation of viral and/or host gene expression to promote favorable conditions, which lead to viral replication [8]. Host induced miRNAs can have anti-viral properties or act as pro-viral factors or help the virus to evade immune response [9]. Studies have indicated that SARS-CoV-2 gains entry to the host cells through adhesion and penetration. Then, after nucleic acid biosynthesis and maturation, viral particles are released from infected cells by exocytosis [10]. During viral infection, a cytokine storm may be induced which leads to an excessive immune response that is detrimental to host cells [11]. Each of these stages is modulated by a subset of miRNAs which can be considered as therapeutic targets in the treatment of COVID-19 patients. Hence, modulation of miRNAs might mitigate COVID-19 pathological negative effects and decrease host damage. Moreover, it has been shown that high-dose single-miRNA administration is needed to achieve highest therapeutic efficiency in vivo, while on the other hand, it can induce adverse effects. Thus, using a cocktail of miRNAs might have reduced off-target side effects and be more efficient than their monotherapy [12], [13]. Here, we review the available data on genome structure and role of immune system during SARS-CoV-2 infection, then functionality of miRNAs in the regulation of this process will be summarized. Finally, we will shed a light on possible therapeutic targets in patients susceptible to severe diseases.
2. SARS-CoV-2 genome structure
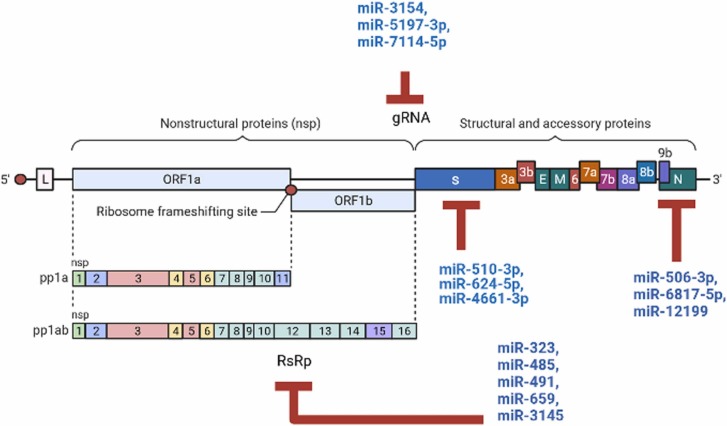
SARS-CoV-2 is an enveloped single stranded positive RNA virus, which possesses a large genome size (29.9 kb), and belongs to the family Coronaviridae in the Nidovirales order and Beta coronavirus (β-CoV) genus [14]. It has been revealed that the CpG dinucleotides in SARS-CoV-2 genome occur with a low frequency that shows virus evolution and adaptation in its hosts [15]. The genome of SARS-CoV-2 comprises a variable number of open reading frames (ORFs) with 5’ and 3’ flanking UTRs which code for accessory proteins, structural proteins including spike (S), membrane (M), and envelope (E) and nucleocapsid (N) along with 16 nonstructural proteins (nsps), which are encoded by the first ORF (ORF1/ab) through a frameshift mutation producing pp1 a and ab ( Fig. 1) [16], [17]. Viral chymotrypsin-like protease (3CLpro) or the main protease (Mpro) and one or two papain-like proteases, play key roles in translational processing of these two polypeptides and convert them into 16 nsps. Nsps are required for replication-transcription complexes (RTC) associated with cellular membranes. Furthermore, ORF3a, ORF6, ORF7a, ORF7b, and ORF8 are processed into six accessory proteins, and structural proteins derived from ORFs 10 and 11 genes [18].
Fig. 1.
Schematic presentation of the SARS-CoV-2 genome structure along with human miRNA-binding sites in the genome.
3. SARS-CoV-2 infection and the immune system
During viral infection, both the innate and adaptive immune responses are activated. Dendritic cells (DCs) and macrophages (Mfs), members of the mononuclear phagocyte system (MPS), are sentinel innate immune cells that combat viruses and bridge the innate and adaptive immune responses by antigen presentation to T cells. Studies have shown that following infection with COVID-19 virus in respiratory system, oxidized phospholipids (OxPLs) are produced through enzymatic or non-enzymatic reactions, which activate monocyte-derived macrophages that lead to induced-production and secretion of type I interferon cytokines such as interleukin 6 (IL-6) and tumor necrosis factor (TNF) [19]. Therefore, type I interferons can induce the expression of angiotensin-converting enzyme 2 (ACE2), a spike protein receptor, which facilities entry of SARS-CoV-2 to the macrophage cytoplasm [20]. Pattern-recognition receptors (PRRs), including toll like receptor (TLR), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) such as retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) at the cytoplasmic level can recognize viral genetic material and induce the innate immune response [21]. For instance, SARS-CoV-2 is recognized by TLR7 in endosomes that is essential in the defense against viral infections, and activates the myeloid differentiating primary response gene 88 (MyD88) pathways and ultimately increases TNFα, IL-1β, IL-6, IL-12, and interferon α (IFNα) expression [22]. IL-8 secreted by cells also can attract neutrophils and T cells which can significantly reduce the number of circulating T cells [23], [24]. TLR3 and TLR4 use TNF receptor associated factor (TRIF) and activate interferon regulatory factor 3 (IFR3) and nuclear factor kappa B (NF-kB), and finally induce production of type I, IFNα/β [25]. Furthermore, TLR4 binds to the spike protein, and also can interact with MyD88 and amplifies the inflammatory responses [25].
In addition, RLRs detect the RNA virus and mainly cooperate in signaling crosstalk networks with TLRs. RIG-I and MDA5 bind to molecule mitochondrial antiviral-signaling protein (MAVS) which leads to activation of the transcription factors, IRF3/7 and NF-κB through recruitment of TRAF3/6 and inhibitory-NF-κB kinase (IKK) [22]. Activation of these transcription factors, in turn, results in upregulation of pro-inflammatory cytokine and IFN production. Based on previous investigations, E, ORF3a, and ORF8b can activate the nod-like receptor protein 3 (NLRP3), a critical multi-protein component of the innate immune system that belongs to the NLR protein family that recruits and activates caspase-1 [26]. Afterwards, pro-IL-1β and IL-18 are cleaved by caspase-1 to their active mature forms (IL-1β and IL-18), which are secreted and induce pyroptosis, a highly inflammatory form of lytic programmed cell death [26], [27], [28]. Furthermore, RIG-I also can induce IL-1β secretion through interaction with apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) independent of NLRP3 [21]. Upregulation and secretion of IL-1β leads to an increased number of activated monocyte-derived macrophages [19]. Furthermore, macrophages secrete IL-12 which increases the activation of natural killer (NK) cells [29]. In turn, type II interferons such as TNF, interferon-γ (IFNγ) and granulocyte–macrophage colony-stimulating factor (GM-CSF) derived from NK cells and T cells create a positive feedback and increase activated macrophages which can initiate tissue damage when overly abundant [19], [29], [30]. In fact, these cytokines activate macrophages through targeting of JAK–STAT and AKT/mTOR/mTORC1 signaling pathways. Studies have also shown that surface receptors of immune cells, Fcγ receptors (FcγRs), can bind Fc portion of IgG and help to internalize IgG bound-virus which results in infection production and more activation of macrophages [19], [31].
Research has also revealed that the body activates cellular and humoral immunity responses against COVID-19 and the majority of cellular immune responses in COVID-19 infection occurs in CD8+ cells [32], [33]. T cells have the strongest immunogenicity to epitopes in the spike, membrane, and nucleoproteins and it has been shown that T helper (Th) cells are needed for effective anti-viral responses by both CD8 T cells and B cells. During viral infections, the antigen-specific immune response is executed by follicular helper CD4 T cells (Tfh) that play a key role in providing assistance to B cells to differentiate and secrete high-affinity antibodies. Differentiated B cells increase the number of germinal center B cell-like (GCB) cells formation in germinal center (GC), the site where activated B cells proliferate, that are required for efficient clearance of the virus [32], [34]. However, in patients with severe COVID diseases reduction in peripheral blood T cells has been reported [35], [36]. In addition, it was found that greater levels of cytokines are secreted in those patients who have more activated and proliferating CD8 and CD4 T cells and Th2 cells. This is associated with a cytokine storm can significantly worsen coronavirus disease [37], [38]. Although T cells are required for proper cellular immune responses to COVID-19 infection, a cytokine storm resulting from an unregulated adaptive immune system can lead to mortality in this disease.
4. The therapeutic role of miRNAs for COVID-19
Research reports have indicated that host miRNAs are actively involved in viral infection and replication. Reciprocally viral miRNAs can make extensive changes in the host transcriptome. Interestingly, it was shown that several miRNAs play vital roles in functionality of various immune cell types and host immune homeostasis. Therefore, due to the importance of miRNAs in the host-pathogen interactions during infection, these small molecules could be considered as anti-viral drug targets. Here, we have shortlisted miRNAs as potential therapeutic targets in COVID-19 ( Table 1).
Table 1.
miRNAs and their targets involved in COVID-19. miRNAs are shown in numerical order.
| miRNA | Changes in expression level | miRNA target | Reference |
|---|---|---|---|
| Replication and pathogenesis | |||
| miR-7–5p, miR-92–3p, miR-98–5p, miR-4500, miR-214–3p, miR-511–3p, miR-6864–3p, let-7e-5p, let-7a-g/i | ↓, ↓, ↓, ↓, ↓, ↓, ↓, ↓, ↓ |
TMPRSS2 | [5], [39], [40], [41] |
| miR-27a/b, miR-106b-5p, miR-130a-3p, miR-141–3p, miR-143, miR-145, miR-200–3p, miR-300, miR-421, miR-429, miR-2113, miR-5197–3p | ↑, ↓, ↓, ↓, ↓, ↑, ↓, ↓, ↓, ↓, ↓, ↓ |
ACE2 | [5], [42], [43], [44], [45] |
| miR-510–3p, miR-624–5p, miR-4661–3p | ↓, ↓, ↓ | S | [46], [47] |
| miR-506–3p, miR-6817–5p, miR-12199 | ↓, ↓, ↓ | N | [9], [48] |
| miR-21–3p | ↑ | replication | [39], [49] |
| miR-323, miR-485, miR-491, miR-654, miR-314 | ↓, ↓, ↓, ↓, ↓ | RdRp | [51] |
| miR-3154, miR-5197–3p, miR-7114–5p | ↓, ↓, ↓ | gRNA | [50], [51] |
| Immune regulatory | |||
| miR-136 | ↑ | RIG-I | [11] |
| miR-21–3p, miR-21–5p, miR-146a, miR-146b, miR-155–5p,miR-200c, let-7 | ↑, ↑, ↓, ↓, ↑, ↓, ↓ |
IL-6 and IL-8 | [11], [52], [53] |
| miR-17 and miR-93–5p | ↓, ↓ | IL-8 | [10] |
| let-7b | ↓ | IFN1 | [54] |
| miR-466i | ↓ | IFNα | [54] |
| miR-5197–3p | ↓ | TGFβ | [50] |
| miR-145 | ↓ | TNFα | [54], [55], [56] |
| miR-146a, miR-351–5p | ↓, ↓ | IL-1β, IL-6, TNFα, IFNα and IFNβ | [54], [55], [56] |
| miR-155 | ↑ | IL-1 | [53] |
| miR-146a | ↓ | IL-1, IL-6 and TNFα | [56] |
| miR-15a/16, miR-21, miR-29a, miR-34a, miRNA-142–5p, miR-146, miR-182, miR-194, miR-1266 | ↓,↑, ↑, ↓, ↑, ↓, ↓, ↓, ↑ |
IL-17 | [57] |
| SARS-CoV-2 coding miRNAs | |||
| miR-147–3p | ↑ | TMPRSS2 | [58], [59] |
| miR-3934–3p, miR-5197, miR-8066 | ↑, ↑, ↑ | S | [58] |
| MD2–5p, miR-147–3p | ↑, ↑ | CHAC1, RAD9 | [60] |
| miR198–3p | ↑ | ADAs | [60] |
| miR-328–5p | ↑ | RXRA | [60] |
| miR-66–3p | ↑ | TNFα | [60] |
| miR-147–5p | ↑ | IFNγ | [60] |
| miR-8066 | ↑ | NF-kB | [58] |
| miR-1468–5p | ↑ | TGF-β1 | [58] |
Note: ↓: decreased expression level; ↑: increased expression level.
5. Therapeutic potential of miRNAs for targeting SARS-CoV-2 replication and pathogenesis
Upon the transmission of virus, SARS-CoV-2 attaches to host-cell receptors and fuses directly with the cell membrane. The SARS-CoV-2 spike protein is cleaved by the transmembrane serine protease 2 (TMPRSS2) located on the host cell membrane and utilizes ACE2 for host cell entry. Therefore, targeting miRNAs, which are involved in expression or activation of TMPRSS2 and/or ACE2 has highly potential for therapeutic applications against COVID-19. As an initial step that can inhibit the host cell entry, miR-98–5p and let-7e-5p can repress TMPRSS2 mRNA in human cells [39], [40]. Other potential miRNAs which have strong binding ability to TMPRSS2 are miR-7–5p, miR-92–3p, miR-214–3p, miR-511–3p, miR-4500, miR-6864–3p and let-7a-g/i [5], [41].
ACE2 is a type 1 integral membrane glycoprotein that serves as transporter of amino acids and viral receptor [42], [43]. A number of studies have suggested that miR-143 and miR-421 are negative regulators of ACE2 while, the level of ACE2 is positively correlated with miR-27a/b and miR-145 expression [5], [44], [45]. Moreover, other potential ACE2 regulators are miR-106b-5p, miR-130a-3p, miR-141–3p, miR-200–3p, miR-300, miR-429, miR-2113 and miR-5197–3p [5], [46], [47]. Therefore, targeting the early attachment of the virus should be considered as an initial and feasible strategy to fight COVID-19.
Khan et al. predicted that host miRNAs can mostly target the ORF1ab and S regions. However, host miRNAs can also bind and target the M, N, ORF3a, ORF7a, ORF8, 5’-UTR, and 3’-UTR regions [9]. Therefore, another potential candidate for an miRNA therapeutic is inhibition of the mRNA encoding the SARS-CoV-2 S protein [48]. In 2020, a study suggested that miR-510–3p and miR-624–5p could interact with the ORF of the viral spike mRNA. However, miR-624–5p suppressed the translation of the spike RNA more efficiently than miR-510–3p [48]. In addition, pab-miRNA-11409d, a miRNA expressed in the gymnosperm Picea abies, that has evolved as a defense against viral infection, also exhibits nucleotide identity to 3' region of SARS-CoV-2 spike mRNA; and thus it can also be considered for COVID-19 treatment [49]. It has been suggested that miR-4661–3p suppresses the S gene of SARS-CoV-2 [50]. Furthermore, targeting nonstructural proteins and the nucleocapsid can also be considered for COVID-19 treatment. For instance, Khan et al., analyzed RNA–RNA interactions of the host miRNA and SARS-CoV-2 genome in 67 patients and showed that miR-506–3p, miR-6817–5p and miR-12199 can interact with the nucleocapsid mRNA of SARS-CoV-2 in most patients [9], [51].
miRNAs which can regulate viral genome replication or regulate their gene expression may also be considered good therapeutic tools. For example, miR-2911, a plant-based miRNA, which is enriched in honeysuckle decoction preparation and efficiently inhibits H5N1 and H7N9 viral replication. This small RNA also has about 30 binding sites on COVID-19 genome, therefore it has been proposed to inhibit the expression of all proteins of this virus and directly suppresses the replication of SARS-CoV-2. Zhou et al., recently showed that exosomes containing miR-2911 significantly block the replication of SARS-CoV-2 [52]. However, miRNA-mediated virus repression is another mechanism to enhance viral replication. For instance, Nersisyan et al., observed that miR-21–3p is 8-fold up-regulated during lung infection by SARS-CoV-2. It has been reported that in the early stages of infection, SARS-CoV-2 interacts with host miR-21–3p to inhibit its replication which leads to a delayed immune response and helps viral survival and improves reproduction [39]. miR-21–3p has a potential binding site on the polyprotein 1a mRNA which is conserved across human coronaviruses. Furthermore, it has been hypothesized that miR-21–3p can also decrease the expression of histone deacetylase 8 (HDAC8) gene, an important modulator of chromatin structure, and therefore inhibits viral replication [39]. RNA-dependent RNA polymerase (RdRp), the RNA polymerase responsible for viral RNA synthesis, is a specific substrate of the virus with no host cellular homologs could be another more effective therapeutic target for treating COVID-19 [53]. To this end, it was shown that host miR-323, miR-485, miR-491, miR-654, and miR-3145 interact with viral RdRp and restrict viral replication [54].
Additionally, use of synthetic complete complementary miRNAs can also be an alternative therapeutic approach for inhibiting the reproduction of coronaviruses. For example, in a bioinformatic analysis of miRNA binding sites on SARS-CoV-2 genome conducted by Ivashchenko et al., it was shown that miR-5197–3p has effective interactions with SARS-CoV-2. However, this miRNA also potentially interacts with the mRNA of few human genes. Therefore, complementary miRNA 2 (cc-miR2) is created by adding G and U nucleotides at the ends of miR-5197–3p to increase the free interaction energy of this cc-miR with the gRNA of SARS-CoV-2. Thus, cc-miR2 specifically interacts with viral gRNA without side effects on human mRNAs [55]. Likewise, Rakhmetullina et al. created some cc-miRs based on miR-3154, miR-5197–3p and miR-7114–5p which could suppress replication of gRNA of SARS-CoV-2 without toxicity or side effects on human coding proteins [56].
Further targeting of the mRNAs of other key proteins such as the Mpro and 3CLpro proteases, with high sequence homology between SARS-CoV and SARS-CoV-2, may be a potential applicable treatment to SARS-CoV-2.
6. Targeting immune regulatory miRNAs
During the cytokine storm, elevated levels of IL-1β, IL-2, IL-6, IL-7, IL-8, IL-9, IL-10, IL-17, G-CSF, GM-CSF, IFNγ, and TNFα, as well as chemokines such as interferon-inducible protein 10 (IP10), monocyte chemoattractant protein 1 (MCP1), macrophage inflammatory protein 1α (MIP1α) and MIP1β have been detected in patients with COVID-19 [57]. Thus, targeting miRNAs which are implicated in exacerbated inflammatory responses will alleviate the complications associated with COVID-19. In mammalian cells, miR-136 binds to the 3′-UTR of RIG-I transcript and promotes IL-6 and IFNβ accumulation. Likewise, the expression levels of IL-6 and IL-8 were significantly increased by miR-21–3p, miR-21–5p and miR-155–5p [11], [58], [59]. On the other hand, miR-146a, miR-146b and miR-200c reduce the expression of IL-6, IL-8, and C-C motif chemokine ligand 5 (CCL5) [60], [61]. Additionally, it has been shown that miR-17 and miR-93–5p suppress the expression of IL-8 [10]. Furthermore, let-7 can regulate TLR4 and subsequently influences the activation of NF-κB. This miRNA also reduces the expression of pro-inflammatory cytokine IL-6 and IL-8 [62], [63]. In addition, let-7b has been shown to regulate type I IFN while IFNα is directly targeted by miR-466i. Bioinformatics analysis also indicated that miR-5197–3p, which is critical for TGFβ pathway, can specifically bind to SARS-CoV-2 gRNA without targeting human genes, and therefore it could have significant therapeutic effects [56]. miR-145 results in modulation of TNFα production whereas, miR-146a or miR-351–5p reduce the production of this pro-inflammatory cytokine [62], [64], [65]. It has been demonstrated that targeting IL-1 and IL-17 may also be useful for blocking cytokine storm and immunopathology in patients with severe COVID-19 [66]. IL-1 is mostly secreted by activated macrophages, neutrophils, and epithelial cells, which enhance the production of T lymphocyte–derived cytokine and Th2 cell responses. miR-155 upregulates IL-1 signaling pathway, while in contrary miR-146a depletion leads to IL-1, IL-6 and TNFα overproduction [59], [64], [67]. Therefore, modulation of these miRNAs would be useful in the control of the immune response in severe COVID-19 patients. IL-17 is a pro-inflammatory cytokine produced by Th17 cells developed from CD4+ T-cells, and excessive production of this cytokine is involved in pathogenesis of several autoimmune diseases. miRNA-142–5p, miR-21, miR-1266, and miR-29a positivity regulate IL-17 whereas miR-146a, miR-182, miR-194, miR-15a/16 and miR-34a can reduce IL-17 levels [68]. Thus, miRNA regulators of IL-17 would provide potential targets for reduction of immune-mediated damage in patients and can alleviate their COVID-19-related immunopathology.
7. Targeting SARS-CoV-2 coding miRNAs
Previous reports have indicated that viral encoded miRNAs target several host immune signaling pathways such as autophagy, IFN and mTOR, etc., and thus, play essential roles in pathology of viral infection [69], [70]. In a study, Liu et al. have predicted that miR-147–3p, which is encoded by SARS-CoV-2 can enhance the expression of TMPRSS2 in the gut and increase the capacity of the virus for transmission [71]. Therefore, suppression of miR-147–3p using an antisense oligonucleotide might have therapeutic benefits. In another study, it was revealed that miR-5197, miR-8066 and miR-3934–3p are involved in N-linked and O-linked glycosylation of subunit S1 and S2 proteins which can increase the pathogenicity of SARS-CoV-2 [72]. Additionally, Yu et al., found three miRNA precursor sequences of the novel coronavirus using their newly developed method which needs further study [73]. Evidence came from the observation that virus-encoded miRNA, MD2–5p and miR-147–3p have regulatory roles on ChaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1) and checkpoint clamp component A (RAD9A) expressions, respectively. Interestingly, CHAC1 is a mammalian pro-apoptotic enzyme downstream of the pancreatic ATF4 eIF2α kinase pathway, while RAD9A is part of a checkpoint protein complex required for DNA damage repair [71]. As it has been reported, miR-198–3p suppresses IFN responses via enhancing the expression of adenosine deaminases acting on RNA (ADARs), which results in prevention of PKR (protein kinase R) and MDA5 activation. Another viral-encoded miRNA, miR-328–5p, suppresses type I interferon immune response through retinoid X receptor alpha (RXRA) activation. However, some miRNAs such as miR-66–3p, miR-147–5p, miR-1468–5p and miR-8066 affect inflammatory responses linked to the cytokine storm [50], [71], [72].
It has been suggested that SARS-CoV-2-encoded miRNAs are involved in apoptosis suppression, regulation of cytoskeleton dynamics and assist viral exocytosis and release. The majority of genes involved in Ca+ signaling are primary targets of viral miRNAs which play important roles as main activators of multiple signaling pathways [9]. Other targets of viral miRNAs are heart and brain development-related pathways, so this viral infection is more severe for the patients with established cardiovascular diseases [9]. Although majority of the miRNAs have a conserved sequence, some mutations have been detected in miR-1307–3p and miR-1468–5p of SARS-CoV-2 strains which may be linked to its pathogenicity [72]. However, it seems that targeting one of highly conserved miRNAs would be an effective approach in prevention and a promising therapeutic strategy for all SARS-CoV-2 strains.
8. Respiratory disease and COVID-19
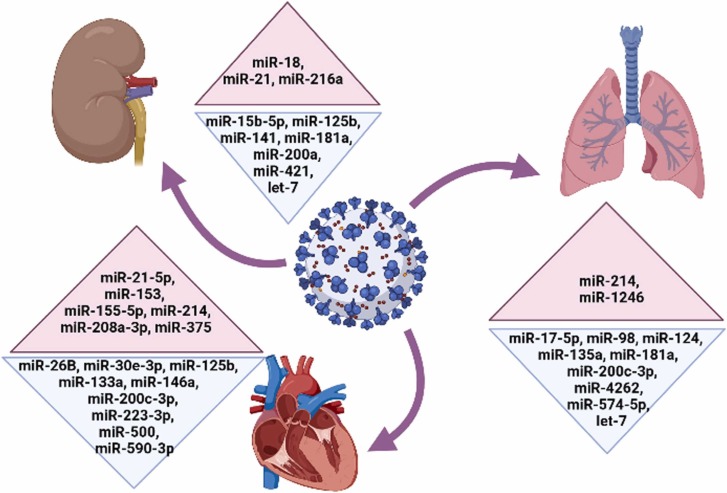
The pulmonary system is the main and first organ system affected by COVID-19. Lung epithelial cells, alveolar macrophages and DCs are the primary cells involved in respiratory immunity [74]. Pulmonary embolism (PE), a serious and potentially life-threatening condition due to a blockage of the pulmonary artery, has been reported in severe COVID-19 patients [75]. Among the identified miRNAs, miR-17–5p is reported to play an anti-viral role in respiratory tract infection and forced expression of this miRNA can be considered as a potential treatment for COVID-19 ( Fig. 2) [76]. ACE2 is highly expressed in lung epithelial cells; therefore, miR-124, miR-135, miR-181a, miR-200c-3p, miR-214, miR-574–5p, miR-1246, miR-4262 and let-7b which regulate the expression of this receptor in respiratory system could be potential candidates for SARS-CoV-2 treatment in lung tissue (Fig. 2). Recent studies have indicated that miR-124 and miR-135a which inhibit the proliferation, migration and invasion of cancer cells induce downregulation of ACE2 protein [1], [77], [78]. Therefore, these miRNAs can be good candidates for treatment of COVID-19 patients with lung cancer. In a study, Cao and colleagues revealed that miR-181a acts as a tumor suppressor miRNA in non-small cell lung cancer (NSCLC) through reduction of IL-17 [79]. miR-181a can indirectly reduce the ACE2 expression via targeting RAS components [80]. Other good approaches for targeting ACE2 protein could be upregulation of miR-200c-3p and suppression of miR-214 which aid in preventing NSCLC [81], [82]. MiR-200c-3p inhibits migration of lung cancer via reduction of HIF1α; while, miR-214 induces tumor proliferation, invasion, and metastasis by FGFR1-WNT/MAPK/AKT signaling pathway [82], [83].
Fig. 2.
Important miRNAs with potential therapeutic benefits for SARS-CoV-2 in patients with respiratory diseases, diabetes, heart failure and kidney diseases. Red and blue colors represent upregulated and downregulated miRNAs in patients, respectively. miRNAs are shown in numerical order.
In pulmonary arterial hypertension (PAH), miR-4262 that affects ACE2 expression indirectly via targeting anti-apoptotic gene BCL2, can also be considered as a promising novel treatment for COVID-19 patients [50], [84]. miR-574–5p is an important negative regulator of pro-inflammatory response that inhibits TLR4/ NF-kB signaling and attenuates acute respiratory distress syndrome (ARDS) [85]. miR-574–5p can also contribute to ACE2 expression as well. Furthermore, in NSCLC cells, overexpression of the oncogenic miR-1246 and suppression of let-7 enhance the tumorigenicity and metastasis [86], [87]. Silencing of miR-1246 alleviates LPS-induced lung inflammation by reduction of IL-1β and TNFα, and inhibition of ACE2 [88]. let-7 directly targets ACE2 and inhibits Idiopathic pulmonary fibrosis (IPF) [89].
In a study, Matares et al. discovered that miR-98 can repress TMPRSS2 mRNA, a cell surface protein primarily expressed by endothelial cells (Fig. 2) [40]. Additionally, this small RNA is significantly less expressed in patients with lung cancer and its modulation leads to suppression of IL-10 in B cells and cancer growth inhibition [90]. Therefore, in patients with respiratory diseases targeting some miRNAs would be a suitable approach for treatment of COVID-19 as it can lead to relieving signs and symptoms of the illness as well as targeting SARS-CoV-2 infection.
9. Impact of diabetes, myocardial damage and heart failure, on COVID-19
Diabetes mellitus (hereafter diabetes) is a metabolic disease in which the body does not produce or respond normally to the insulin; thus, it cannot regulate blood glucose levels. Studies have found a relationship between diabetes and increased severity and mortality of coronavirus disease [91], [92], [93]. Several studies have observed increased cellular adaptive stress and thrombotic tendency due to insulin inactivity that may lead to vascular complications and heart failure which can be intensified by lung dysfunction after COVID-19 infection [91], [94], [95], [96], [97], [98], [99]. Nevertheless, modulation of miRNAs on cardiovascular system can serve as a therapeutic approach for patients with diabetes and COVID-19.
Some reports suggest that there are higher risks of SARS-CoV-2 infection, severe illness or mortality among individuals with underlying cardiovascular diseases. Coronaviruses are known to induce damage in cardiovascular system in several ways [100], [101], [102]. It has been shown that infection of cardiomyocytes with SARS-CoV-2 can change the expression of genes involved in cellular metabolism and immune response, and induce cytotoxic effects. Increases in pro-inflammatory cytokines affect heart arrhythmias, rapid blood hypotension, restricted blood flow and coagulation with microvascular clotting causing occlusion of small vessels. Anti-viral drugs also can increase cardiac dysfunction. Therefore, managing cardiovascular conditions in COVID-19 patients appears to be essential.
Elevated miR-153, miR-208a-3p and miR-375 levels are associated with myocardial damage [3], [103], [104], therefore downregulation of these miRNAs could reduce severe COVID-19 infection in patients with pre-existing cardiovascular diseases (CVDs) (Fig. 2). miR-208a and miR-375, both of which are highly expressed in dilated cardiomyopathy (DCM) patients, can induce apoptosis in ischemic cardiomyocytes and their downregulation improves cardiac function [3], [103], [104].
ACE2 receptors are widely expressed in the vascular endothelium and cardiomyocytes that are involved in heart function and diabetes mellitus [2], [3], [105], [106]. Thus, targeting ACE2 expression using miRNAs like miR-26b-5p and miR-200c-3p can be important for COVID-19 infection in patients with pre-existing CVDs [47], [107], [108].
It has been demonstrated that miR-21–5p, miR-155–5p and miR-214 which are overexpressed in heart failure, control the release of pro-inflammatory cytokines in the heart, while miR-125b and miR-223–3p improve cardiac function and target components of the insulin signaling pathway. Thus, these mRNAs effectively reduce high glucose induced endothelial cell injury (Fig. 2) [3], [109], [110], [111], [112], [113]. In addition, these miRNAs also play an anti-inflammatory role [112], [114]. Therefore, modulation of these miRNAs can be considered as a therapeutic approach for patients with diabetes/CVD and COVID-19. Likewise, dysregulation of miR-590–3p involved in enriched IL-6 and TNFα expression can lead to myocarditis and heart dysfunction. Thus, upregulation of this small RNA effectively restores the cardiac function by suppressing NF-κB [112]. miR-146a can attenuate myocardial injury and inhibit inflammation through reduction of NF-κB level [115]. Furthermore, this miRNA is significantly dysregulated in diabetes [64], thus forced expression of miR-146a may decreased the poor clinical outcome of COVID-19 in patients with sepsis induced myocardial dysfunction or diabetes (Fig. 2). There exists some evidence that miR-133a abundance is significantly downregulated in the hearts of diabetic patients which causes an upregulation of serum and glucocorticoid regulated kinase 1 (SGK1) and insulin-like growth factor receptor 1 (IGFR1) and finally leads to cardiac hypertrophy [116]. miR-133a also has anti-inflammatory function through targeting IL-6 and IL-8 expression [11]. It appears that upregulation of this miRNA could reduce the severity of COVID-19 in patients with diabetes (Fig. 2). Another miRNA involved in myocardial injury that can target SARS-CoV-2 is miR-30e-3p. It is speculated that this miRNA may suppress the replication of the virus through binding to a complementary site on the viral RNA genome [2]. Taken together, application of anti-miRNAs which improve cardiac function along with modulation of specific miRNAs against COVID-19 would help to relieve symptoms in patients with severe coronavirus disease.
10. Kidney disease and COVID-19
There are some reports demonstrating that renal diseases substantially increase the risk of SARS-CoV-2 infection, and severity and mortality of coronavirus disease [117], [118]. Since the only other organ other than the heart with elevated ACE2 expression is the kidney, it can be a target of COVID-19 infection [119], [120]. Interestingly, some previous studies have shown adverse effects of SARS-CoV-2 on kidney which causes podocyte injury and protein loss through the urine [121], [122], [123].
The consequence of SARS-CoV-2 on kidney is dehydration which leads to a decline in glomerular filtration rate (GFR) and acute kidney injury. Another possible mechanism of viral injury is sepsis that can lead to a cytokine storm through innate immune response. Rhabdomyolysis (RM), a rare but serious muscular condition, has been reported in COVID-19 patients, which can lead to kidney damage [124]. Further potential mechanisms that SARS-CoV-2 can result in injury on kidney are hypertension and toxic drugs. Previous reports indicate that loss of miRNA-141, miRNA-200a and let-7 along with upregulation of miR-216a increase kidney fibrosis via overexpression of TGFβ expression [125], [126]. Therefore, manipulating the expression of these miRNAs could lead to protective effects against kidney fibrosis and COVID-19 severity through inhibition of TGFβ signaling (Fig. 2).
It has also been revealed that downregulation of miR-15b-5p is involved in sepsis-induced acute kidney injury (AKI) via mTOR signaling pathway [127], [128]. Kim and colleagues in 2020 showed that this miRNA can bind to SARS-CoV-2 (Fig. 2) [129]. Therefore, more studies are needed to investigate therapeutic potentials of miR-15b-5p.
Regarding the importance of ACE2 related changes in renal injury during SARS-CoV-2 infection, targeting miRNAs that regulate ACE2 network would be useful in COVID patients with chronic kidney disease as well. For example, it is experimentally validated that miR-181a can target the ACE2 mRNA (Fig. 2) [84]. Furthermore, there is a negative correlation between miR-181a and hypertension association with renin. It has been evident that upregulation of miR-181a is associated with upregulation of signaling cascades of adaptive immunity and inflammation [130]. Thus, forced expression of this miR can serve as a therapeutic agent for COVID-19 [131].
Another miRNA that can specifically be targeted for curing COVID-19 in patients with chronic kidney disease is miR-421 (Fig. 2) [131]. Trojanowicz et al., have shown that miR-421 binds to 3'-UTR of the ACE2 mRNA [132]. A study conducted by Huanghas revealed that miR-125b is required for ACE2 reduction upon high glucose exposure in HK-2 renal tubular epithelial cells; therefore it can be a good candidate for prevention of diabetic nephropathy (Fig. 2) [57]. miR-18 which is highly upregulated following kidney injury and nephropathy, also plays a crucial role on ACE2 expression by upregulating the NOX2/ROS pathway [133]. Thus, downregulating ACE2 via an anti-miR-18 could potentially pave the way for COVID-19 treatment (Fig. 2). Overexpression of miR-21 which was found to be related over-activated immune response and viral replication, can also lead to renal injury [39], [125], [126]. Thus, miR-21 knockdown is suggested to serve as a potential therapeutic agent against SARS-CoV-2 (Fig. 2). Taken together, harnessing miRNAs which suppress ACE2 along with modulation of immune response might have great curative potential against COVID-19 associated nephropathy.
11. Conclusion and future perspectives
COVID-19 is an infectious and highly transmittable disease which continues to represent a most formidable challenge to medicine and public health [134]. Considering that the elderly and those with underlying medical conditions such as lung disease, diabetes, heart failure or kidney diseases are at increased risk for severe COVID-19, therefore, effective, novel and targeted treatments are required against the cause of this infectious disease [1], [2], [3], [4].
A growing number of studies have shown that host and viral encoded ncRNAs play important roles in pathology and pathogenesis of human viral infections [8], [9]. Therefore, targeting the expression of miRNAs results in modification of various signaling pathways and cellular processes, which can affect the therapeutic efficacy against viral infections. In this regard, finding specific miRNAs with potential therapeutic benefits against SARS-CoV-2 depends on the patient's particular condition or illness, could be considered as a promising strategy to reduce COVID-19 severity. For instance, regarding to the importance of ACE2 in SARS-CoV-2 infection, miRNA modulators of the ACE2 network might be helpful as therapeutic targets for personalized treatment of COVID-19 patients. Likewise, other treatment candidates that target miRNAs involved in the immune response in order to calm the cytokine storm and other aggressive inflammatory response in COVID-19 patients.
Combined treatment with anti-inflammatory and anti-viral miRNAs could be more effective than either of them alone. Meanwhile, targeting viral genome, key proteins or miRNAs could be a potential antiviral strategy for developing effective treatments against this disease. Since targeting a single miRNA may regulate a large number of target mRNAs, high-dosage of a single miRNA in vivo can lead to serious off-target negative side effects. Thus, using a system biology approach for synergistic treatments could require lower doses of a cocktail of multiple therapeutic key microRNAs may effectively clear SARS-CoV-2 without any adverse effects.
Generally, targeted delivery, instability and toxicity are the main challenges of miRNA-based therapy [135], therefore suitable carrier vehicles are needed for efficient and safe delivery of miRNAs to the site of infection. In this regard, studies have shown that mesenchymal stem cells (MSCs) exhibit tropism for sites of injury or inflammation [136], [137] and can potentially be used to attenuate the cytokine storm. Thus, use of MSCs or their cell-free products might be considered as a promising treatment option and suitable carriers for delivery of miRNAs to the sites of SARS-CoV-2 infection. Cumulatively, targeting a number of key miRNAs might be an effective and highly promising tool against the virus; however, further research is required to candidate specific miRNAs before they can reach clinical trials.
CRediT authorship contribution statement
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, writing, or revision of the manuscript. N.A.: Manuscript writing, revision of the manuscript;. T.N.: Financial support, manuscript writing, revision of the manuscript;. M. MM.: Manuscript writing, revision of the manuscript.
Conflict of interest statement
The authors declare that they have no competing interests.
Acknowledgments
TN would like to acknowledge financial support from Vetenskapsrådet (The Swedish Research Council, 2017-04663) and Carl Tryggers Stiftelse (CTS:18:279).
References
- 1.Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Noncoding RNA Res. 2020;5(4):153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra P.K., Tandon RByrareddy S.N. Diabetes and COVID-19 risk: an miRNA perspective. Am. J. Physiol. Heart Circ. Physiol. 2020;319(3):604. doi: 10.1152/ajpheart.00489.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greco S., Madè A., Gaetano C., Devaux Y., Emanueli C., Martelli F. Noncoding RNAs implication in cardiovascular diseases in the COVID-19 era. J. Transl. Med. 2020;18(1):408. doi: 10.1186/s12967-020-02582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Wang J., Xu Y., Guo M., Mi K., Xu R., Pei Y., Zhang Q., Luan X., Hu Z. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int. J. Mol. Sci. 2020;21:4807. doi: 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhopadhyay D., Mussa B.M. Identification of novel hypothalamic microRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in Silico analysis. Brain Sci. 2020;10(10):666. doi: 10.3390/brainsci10100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardini B., Calin G.A. MicroRNAs and Long non-coding RNAs and their hormone-like activities in cancer. Cancers. 2019;11(3):378. doi: 10.3390/cancers11030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundhoff A., Sullivan C.S. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan M.A., Sany M.R.U., Islam MSIslam A. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasparello J., Finotti A., Gambari R. Tackling the COVID-19 “cytokine storm” with microRNA mimics directly targeting the 3’UTR of pro-inflammatory mRNAs. Med. Hypotheses. 2021;146 doi: 10.1016/j.mehy.2020.110415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai X., Eberhardt M., Schmitz U., Vera J. Systems biology-based investigation of cooperating microRNAs as monotherapy or adjuvant therapy in cancer. Nucleic Acids Res. 2019;47(15):7753–7766. doi: 10.1093/nar/gkz638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cursons J., Pillman K.A., Scheer K.G., Gregory P.A., Foroutan M., Hediyeh-Zadeh S., Toubia J., Crampin E.J., Goodall G.J., Bracken C.P., Davis M.J. Combinatorial targeting by microRNAs co-ordinates posttranscriptional control of EMT. Cell Syst. 2018;7(1):77–91. doi: 10.1016/j.cels.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 14.Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARSCoV-2): an update. Cureus. 2020;12(3):7423. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and Bat RaTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts. Genes. 2020;11(7):761. doi: 10.3390/genes11070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 17.Ying H., Ebrahimi M., Keivan M., Khoshnam S.E., Salahi S., Farzaneh M. MiRNAs; a novel strategy for the treatment of COVID-19. Cell Biol. Int. 2021;45:2045–2053. doi: 10.1002/cbin.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9(4):1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116(6):1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal Ms S.N., Akmal W., Sultan R., Abdullah H., Qindeel M., Dhama K., Bila M. Role of toll-like receptors in coronavirus infection and immune response. J. Exp. Biol. Agric. Sci. 2020;8:S66–S78. [Google Scholar]
- 26.Kelley N., Jeltema D., Duan Y., He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019;20(13):3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink S.L., Cookson B.T. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8(11):1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 28.He W.T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.H., Zhong C.Q., Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell. Res. 2015;25(12):1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parihar R., Dierksheide J., Hu Y., Carson W.E. IL-12 enhances the natural killer cell cytokine response to Ab-coated tumor cells. J. Clin. Invest. 2002;110(7):983–992. doi: 10.1172/JCI15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y., Liu C.H., Roberts A.I., Das J., Xu G., Ren G., Zhang Y., Zhang L., Yuan Z.R., Tan H.S., Das G. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell. Res. 2006;16(2):126–133. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 31.Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20(10):633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X., Dai T., Zhou X., Qian H., Guo R., Lei L., Zhang X., Zhang D., Shi L., Cheng Y., Hu J. Naturally activated adaptive immunity in COVID-19 patients. J. Cell Mol. Med. 2020;24(21):12457–12463. doi: 10.1111/jcmm.15771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., Dong L. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U.S.A. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, china. Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M., Douek D.C. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181(8):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer K., Harris T. An effective COVID-19 vaccine needs to engage T cells. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.581807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nersisyan S., Engibaryan N., Gorbonos A., Kirdey K., Makhonin A., Tonevitsky A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ. 2020;8:9994. doi: 10.7717/peerj.9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matarese A., Gambardella J., Sardu C., Santulli G. MiR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. 2020;8(11):462. doi: 10.3390/biomedicines8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fani M., Zandi M., Ebrahimi S., Soltani SAbbasi S. The role of miRNAs in COVID-19 disease. Future Virol. 2021;16:301–306. doi: 10.2217/fvl-2020-0389. [DOI] [Google Scholar]
- 42.Yang J., Petitjean S.J., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11(1):4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hum C., Loiselle J., Ahmed N., Shaw T.A., Toudic C., Pezacki J.P. MicroRNA mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Drugs. 2021;81(5):517–531. doi: 10.1007/s40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L.J., Xu R., Yu H.M., Chang Q., Zhong J.C. The ACE2/Apelin signaling, microRNAs, and hypertension. Int J. Hypertens. 2015;2015 doi: 10.1155/2015/896861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert D.W., Lambert L.A., Clarke N.E., Hooper N.M., Porter K.E., Turner A.J. Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421. Clin. Sci. 2014;127(4):243–249. doi: 10.1042/CS20130420. [DOI] [PubMed] [Google Scholar]
- 46.Niu W., Wu F., Cui H., Cao W., Chao Y., Wu Z., Fan M., Liang C. Network pharmacology analysis to identify phytochemicals in traditional chinese medicines that may regulate ACE2 for the treatment of COVID-19. eCAM. 2020;2020:1–14. doi: 10.1155/2020/7493281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., Bär C., Thum T. MicroRNAs targeting the SARSCoV-2 entry receptor ACE2 in cardiomyocytes. JMCC. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallicano G.I., Casey J.L., Fu J., Mahapatra S. Molecular targeting of vulnerable RNA sequences in SARS CoV-2: identifying clinical feasibility. Gene Ther. 2020:1–8. doi: 10.1038/s41434-020-00210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chauhan N., Jaggi M., Chauhan S.C., Yallapu M.M. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti Infect. Ther. 2021;19(2):137–145. doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao H., Gao F., Xie G., Liu Z. Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary endothelial cells during acute lung injury through suppressing miR-4262. Cell Physiol. Biochem. 2015;37(2):759–767. doi: 10.1159/000430393. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed S.S., Paramasivam P., Raj K., Kumar V., Murugesan R., Ramakrishnan V. Regulatory cross talk between SARS-CoV-2 receptor binding and replication machinery in the human host. Front. Physiol. 2020;11:802. doi: 10.3389/fphys.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L.K., Zhou Z., Jiang X.M., Zheng Y., Chen X., Fu Z., Xiao G., Zhang C.Y., Zhang L.K., Yi Y. Absorbed plant miR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6(54) doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23(1):80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivashchenko A.T., Rakhmetullina A.K., Aisina D.E., Akimniyazova A.N. How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV. Res. Sq. 2020:1–13. doi: 10.21203/rs.3.rs-16264/v1. [DOI] [Google Scholar]
- 56.Rakhmetullina A., Ivashchenko A., Akimniyazova A., Aisina D., Pyrkova A. The miRNA complexes against coronaviruses COVID-19, SARS-CoV, and MERS-CoV. Res. Sq. 2020:1–16. doi: 10.21203/rs.3.rs-19592/v1. [DOI] [Google Scholar]
- 57.Huang Y.F., Zhang Y., Liu C.X., Huang J., Ding G.H. MicroRNA-125b contributes to high glucoseinduced reactive oxygen species generation and apoptosis in HK-2 renal tubular epithelial cells by targeting angiotensin-converting enzyme 2. Eur. Rev. Med. Pharm. Sci. 2016;20(19):4055–4062. [PubMed] [Google Scholar]
- 58.Pfeiffer D., Roßmanith E., Lang I., Falkenhagen D. MiR-146a, miR-146b, and miR-155 increase expression of IL-6 and IL-8 and support HSP10 in an in vitro sepsis model. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soni D.K., Cabrera-Luque J., Kar S., Sen C., Devaney J., Biswas R. Suppression of miR-155 attenuates lung cytokine storm induced by SARS-CoV-2 infection in human ACE2-transgenic mice. bioRxiv. 2020:1–29. doi: 10.1101/2020.12.17.423130. [DOI] [Google Scholar]
- 60.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. The miRNA: a small but powerful RNA for COVID-19. Brief. Bioinform. 2021;22(2):1137–1149. doi: 10.1093/bib/bbab062. 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abu-Izneid T., AlHajri N., Ibrahim A.M., Javed M.N., Salem K.M., Pottoo F.H., Kamal M.A. Micro-RNAs in the regulation of immune response against SARS COV-2 and other viral infections. J. Adv. Res. 2020;30:133–135. doi: 10.1016/j.jare.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu H.R., Hsu T.Y., Huang H.C., Kuo H.C., Li S.C., Yang K.D., Hsieh K.S. Comparison of the functional microRNA expression in immune cell subsets of neonates and adults. Front. Immunol. 2016;7:615. doi: 10.3389/fimmu.2016.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mormile R. Diabetes and susceptibility to COVID-19: may miR-146a make the difference between life and death? Minerva Endocrinol. 2021 doi: 10.23736/S2724-6507.21.03395-2. [DOI] [PubMed] [Google Scholar]
- 65.Gambardella J., Sardu C., Morelli M.B., Messina V., Castellanos V., Marfella R., Maggi P., Paolisso G., Wang X., Santulli G. Exosomal microRNAs drive thrombosis in COVID-19. medRxiv. 2020:1–7. doi: 10.1101/2020.06.16.20133256. [DOI] [Google Scholar]
- 66.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Testa U., Pelosi E., Castelli G., Labbaye C. MiR-146 and miR-155: two key modulators of immune response and tumor development. Noncoding RNA. 2017;3(3):22. doi: 10.3390/ncrna3030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mai J., Virtue A., Maley E., Tran T., Yin Y., Meng S., Pansuria M., Jiang X., Wang H., Yang X.F. MicroRNAs and other mechanisms regulate interleukin-17 cytokines and receptors. Front Biosci. 2012;4:1478–1495. doi: 10.2741/474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y., Geng P., Liu Y., Wu J., Qiao H., Xie Y., Yin N., Chen L., Lin X., Liu Y., Yi S. Rotavirus-encoded virus-like small RNA triggers autophagy by targeting IGF1R via the PI3K/Akt/mTOR pathway. Biochim. Biophys. Acta BBA Molecul. Basis Dis. 2018;1864(1):60–68. doi: 10.1016/j.bbadis.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 70.Ahmad I., Valverde A., Siddiqui H., Schaller S., Naqvi A.R. Viral microRNAs: interfering the interferon signaling. Curr. Pharm. Des. 2020;26(4):446–454. doi: 10.2174/1381612826666200109181238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z., Wang J., Xu Y., Guo M., Mi K., Xu R., Pei Y., Zhang Q., Luan X., Hu Z. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int. J. Mol. Sci. 2020;21:1–24. doi: 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., Uysal-Onganer P. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu T.Y., Chen M., Wang C.D. Annotation of miRNAs in the COVID-19 novel coronavirus. J. Electron. Sci. Technol. 2021;19(1):1–8. doi: 10.1016/j.jnlest.2020.100060. [DOI] [Google Scholar]
- 74.Bissonnette E.Y., Lauzon-Joset J.F., Debley J.S., Ziegler S.F. Cross-talk between alveolar macrophages and lung epithelial cells is essential to maintain lung homeostasis. Front. Immunol. 2020;11:1–12. doi: 10.3389/fimmu.2020.583042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakr Y., Giovini M., Leone M., Pizzilli G., Kortgen A., Bauer M., Tonetti T., Duclos G., Zieleskiewicz L., Buschbeck S., Ranieri V.M. Pulmonary embolism in patients with coronavirus disease-2019 (COVID-19) pneumonia: a narrative review. Ann. Intensive Care. 2020;10(1):1–3. doi: 10.1186/s13613–020-00741–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sardar R., Satish D., Gupta D. Identification of novel SARS-CoV-2 drug targets by host microRNAs and transcription factors co-regulatory interaction network analysis. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.571274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang M., Meng B., Liu Y., Yu J., Chen Q. MiR-124 inhibits growth and enhances radiation-induced apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell Physiol. Biochem. 2017;44(5):2017–2028. doi: 10.1159/000485907. [DOI] [PubMed] [Google Scholar]
- 78.Tian Y., Zhang L., Yu Q., Wang Z., Yang X. MiR-135a inhibits non-small cell lung cancer progression by suppressing RAB1B expression and the RAS pathway. Aging. 2020;12(14):14480–14489. doi: 10.18632/aging.103494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao Y., Zhao D., Li P., Wang L., Qiao B., Qin X., Li L., Wang Y. MicroRNA-181a-5p impedes IL-17- induced nonsmall cell lung cancer proliferation and migration through targeting VCAM-1. Cell Physiol. Biochem. 2017;42(1):346–356. doi: 10.1159/000477389. [DOI] [PubMed] [Google Scholar]
- 80.Marques F.Z., Campain A.E., Tomaszewski M., Zukowska-Szczechowska E., Yang Y.H., Charchar F.J., Morris B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58(6):1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 81.Li J., Tan Q., Yan M., Liu L., Lin H., Zhao F., Bao G., Kong H., Ge C., Zhang F., Yu T. MiRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol. Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y., Li Z., Yuan H., Ji W., Wang K., Lu T., Yu Y., Zeng Q., Li F., Xia W., Lu S. Reciprocal regulatory mechanism between miR-214-3p and FGFR1 in FGFR1-amplified lung cancer. Oncogenesis. 2019;8(9):50. doi: 10.1038/s41389-019-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Byun Y., Choi Y.C., Jeong Y., Lee G., Yoon S., Jeong Y., Yoon J., Baek K. MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells. Cell. Mol. Biol. Lett. 2019;24(28):28. doi: 10.1186/s11658-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Badawi S., Ali B.R. ACE2 nascence, trafficking, and SARS-CoV-2 pathogenesis: the saga continues. Hum. Genom. 2021;15(1):8. doi: 10.1186/s40246-021-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He B., Zhou W., Rui Y., Liu L., Chen B., Su X. MicroRNA-574-5p attenuates acute respiratory distress syndrome by targeting HMGB1. Am. J. Respir. Cell. Mol. Biol. 2021;64(2):196–207. doi: 10.1165/rcmb.2020-0112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Du P., Lai Y.H., Yao D.S., Chen J.Y., Ding N. Downregulation of microRNA-1246 inhibits tumor growth and promotes apoptosis of cervical cancer cells by targeting thrombospondin-2. Oncol. Lett. 2019;18(3):2491–2499. doi: 10.3892/ol.2019.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tristán-Ramos P., Rubio-Roldan A., Peris G., Sánchez L., Amador-Cubero S., Viollet S., Cristofari G., Heras S.R. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat. Commun. 2020;11(1):5712. doi: 10.1038/s41467-020-19430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fang Y., Gao F., Hao J., Liu Z. MicroRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am. J. Transl. Res. 2017;9(3):1287–1296. [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang R., Su H., Ma X., Xu X., Liang L., Ma G., Shi L. MiRNA let-7b promotes the development of hypoxic pulmonary hypertension by targeting ACE2. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;316(3):547. doi: 10.1152/ajplung.00387.2018. [DOI] [PubMed] [Google Scholar]
- 90.Li Y., Rong J., Qin J., He J.Y., Chen H.G., Huang S.H. Micro RNA-98 interferes with expression interleukin-10 in peripheral B cells of patients with lung cancer. Sci. Rep. 2016;6:32754. doi: 10.1038/srep32754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lim S., Bae J.H., Kwon H.S., Nauck M.A. COVID-19 and diabetes mellitus: from pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021;17:11–30. doi: 10.1038/s41574-020-00435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muniyappa R., Gubbi S. COVID-19 pandemic, coronaviruses, and diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 2020;318(5):E736–E741. doi: 10.1152/ajpendo.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ugwueze C.V., Ezeokpo B.C., Nnolim B.I., Agim E.A., Anikpo N.C., Onyekachi K.E. COVID-19 and diabetes mellitus: the link and clinical implications. Int. J. Diabetes Metab. 2020;26(2):69–77. doi: 10.1159/000511354. [DOI] [Google Scholar]
- 94.Colwell J.A. Vascular thrombosis in type II diabetes mellitus. Diabetes. 1993;42(1):8–12. doi: 10.2337/diab.42.1.8. [DOI] [PubMed] [Google Scholar]
- 95.Calabrese V., Cornelius C., Leso V., Trovato-Salinaro A., Ventimiglia B., Cavallaro M., Scuto M., Rizza S., Zanoli L., Neri S., Castellino P. Oxidative stress, glutathione status, sirtuin and cellular stress response in type 2 diabetes. Biochim Biophys. Acta Mol. Basis Dis. 2012;1822(5):729–736. doi: 10.1016/j.bbadis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 96.Donnelly R., Emslie-Smith A.M., Gardner I.D., Morris A.D. Vascular complications of diabetes. BMJ. 2000;320(7241):1062–1066. doi: 10.1136/bmj.320.7241.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsia C.C., Raskin P. Lung function changes related to diabetes mellitus. Diabetes Technol. Ther. 2007;9 Suppl 1(S1):73–82. doi: 10.1089/dia.2007.0227. [DOI] [PubMed] [Google Scholar]
- 98.Tiengo A., Fadini G.P., Avogaro A. The metabolic syndrome, diabetes and lung dysfunction. Diabetes Metab. 2008;34(5):447–454. doi: 10.1016/j.diabet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 99.Yaribeygi H., Sathyapalan T., Jamialahmadi T., Sahebkar A. The impact of diabetes mellitus in COVID-19: a mechanistic review of molecular interactions. J. Diabetes Res. 2020;2020 doi: 10.1155/2020/5436832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bikdeli B., Madhavan M.V., Jimenez D., Chuich T., Dreyfus I., Driggin E., Nigoghossian C.D., Ageno W., Madjid M., Guo Y., Tang L.V. COVID-19 and thrombotic or thromboembolic disease: implications for prevention, antithrombotic therapy, and follow-up: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2020;75(23):2950–2973. doi: 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soumya R.S., Unni T.G., Raghu K.G. Impact of COVID-19 on the cardiovascular system: a review of available reports. Cardiovasc. Drugs Ther. 2021;35:411–425. doi: 10.1007/s10557-020-07073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun T., Dong Y.H., Du W., Shi C.Y., Wang K., Tariq M.A., Wang J.X., Li P.F. The role of microRNAs in myocardial infarction: from molecular mechanism to clinical application. Int. J. Mol. Sci. 2017;18(4):745. doi: 10.3390/ijms18040745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garikipati V.N., Verma S.K., Jolardarashi D., Cheng Z., Ibetti J., Cimini M., Tang Y., Khan M., Yue Y., Benedict C., Nickoloff E. Therapeutic inhibition of miR-375 attenuates post-myocardial infarction inflammatory response and left ventricular dysfunction via PDK-1-AKT signalling axis. Cardiovasc. Res. 2017;113(8):938–949. doi: 10.1093/cvr/cvx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizuiri S., Hemmi H., Arita M., Ohashi Y., Tanaka Y., Miyagi M., Sakai K., Ishikawa Y., Shibuya K., Hase H., Aikawa A. Expression of ACE and ACE2 in individuals with diabetic kidney disease and healthy controls. AJKD. 2008;51(4):613–623. doi: 10.1053/j.ajkd.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 106.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116(6):1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Magenta A., Cencioni C., Fasanaro P., Zaccagnini G., Greco S., Sarra-Ferraris G., Antonini A., Martelli F., Capogrossi M.C. MiR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011;18(10):1628–1639. doi: 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wicik Z., Eyileten C., Jakubik D., Simões S.N., Martins D.C., Pavão R., Siller-Matula J.M., Postula M. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 2020;9(11):3743. doi: 10.3390/jcm9113743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das A., Ganesh K., Khanna S., Sen C.K., Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 2014;192(3):1120–1129. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma X., Becker Buscaglia L.E., Barker J.R., Li Y. MicroRNAs in NF-κB signaling. J. Mol. Cell Biol. 2011;3(3):159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deng B., Hu Y., Sheng X., Zeng H., Huo Y. MiR- 223- 3p reduces high glucose and high fat- induced endothelial cell injury in diabetic mice by regulating NLRP3 expression. Exp. Ther. Med. 2020;20(2):1514–1520. doi: 10.3892/etm.2020.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mirna M., Paar V., Rezar R., Topf A., Eber M., Hoppe U.C., Lichtenauer M., Jung C. MicroRNAs in inflammatory heart diseases and sepsis-induced cardiac dysfunction: a potential scope for the future? Cells. 2019;8(11):1352. doi: 10.3390/cells8111352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du X., Li X., Chen L., Zhang M., Lei L., Gao W., Shi Z., Dong Y., Wang Z., Li X., Liu G. Hepatic miR-125b inhibits insulin signaling pathway by targeting PIK3CD. J. Cell. Physiol. 2018;233(8):6052–6066. doi: 10.1002/jcp.26442. [DOI] [PubMed] [Google Scholar]
- 114.Wang F., Chen X., Song Y., Huang S., Zhou C., Huang C., Chen Z., Zhang L., Ji Y. MiR-223-3p suppresses inflammation to protect cardiomyocytes by targeting NLRP3 in acute myocardial infarction patients. Food Sci. Technol. 2020:1–8. doi: 10.1590/fst.25020. [DOI] [Google Scholar]
- 115.An R., Feng J., Xi C., Xu J., Sun L. MiR-146a attenuates sepsis-induced myocardial dysfunction by suppressing IRAK1 and TRAF6 via targeting ErbB4 expression. Oxid. Med. Cell. Longev. 2018;2018:1–10. doi: 10.1155/2018/7163057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shen E., Diao X., Wang X., Chen R., Hu B. MicroRNAs involved in the mitogen-activated protein kinase cascades pathway during glucose-induced cardiomyocyte hypertrophy. Am. J. Clin. Pathol. 2011;179(2):639–650. doi: 10.1016/j.ajpath.2011.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y., Lv Y., Liu Q. SARS-CoV-2 infection associated acute kidney injury in patients with preexisting chronic renal disease: a report of two cases. Immun. Inflamm. Dis. 2020;8(4):506–511. doi: 10.1002/iid3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31(8):1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mizuiri SOhashi Y. ACE and ACE2 in kidney disease. World J. Nephrol. 2015;4(1):74–82. doi: 10.5527/wjn.v4.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirsch J.S., Ng J.H., Ross D.W., Sharma P., Shah H.H., Barnett R.L., Hazzan A.D., Fishbane S., Jhaveri K.D., Abate M., Andrade H.P. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020;98(1):209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peleg Y., Kudose S., D’Agati V., Siddall E., Ahmad S., Nickolas T., Kisselev S., Gharavi A., Canetta P. Acute kidney injury due to collapsing glomerulopathy following COVID-19 infection. Kidney Int. Rep. 2020;5(6):940–945. doi: 10.1016/j.ekir.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98(1):219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chueh T.I., Zheng C.M., Hou Y.C., Lu K.C. Novel evidence of acute kidney injury in COVID-19. J. Clin. Med. 2020;9(11):3547. doi: 10.3390/jcm9113547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kellum J.A., Nadim M.K., Forni L.G. Sepsis-associated acute kidney injury: is COVID-19 different? Kidney Int. 2020;98(6):1370–1372. doi: 10.1016/j.kint.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu H., Kong L., Zhou S., Cui W., Xu F., Luo M., Li X., Tan Y., Miao L. The role of microRNAs in diabetic nephropathy. J. Diabetes Res. 2014;2014 doi: 10.1155/2014/920134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cao Q., Chen X.M., Huang C., Pollock C.A. MicroRNA as novel biomarkers and therapeutic targets in diabetic kidney disease: an update. FASEB BioAdv. 2019;1(6):375–388. doi: 10.1096/fba.2018-00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu G., Mo L., Wu C., Shen X., Dong H., Yu L., Pan P., Pan K. The miR-15a-5p-XIST-CUL3 regulatory axis is important for sepsis-induced acute kidney injury. Ren. Fail. 2019;41(1):955–966. doi: 10.1080/0886022X.2019.1669460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fan P.C., Chen C.C., Chen Y.C., Chang Y.S., Chu P.H. MicroRNAs in acute kidney injury. Hum. Genom. 2016;10(1):29. doi: 10.1186/s40246-016-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim W.R., Park E.G., Kang K.W., Lee S.M., Kim B., Kim H.S. Expression analyses of microRNAs in hamster lung tissues infected by SARS-CoV-2. Mol. Cells. 2020;43(11):953–963. doi: 10.14348/molcells.2020.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marques F.Z., Romaine S.P., Denniff M., Eales J., Dormer J., Garrelds I.M., Wojnar L., Musialik K., DudaRaszewska B., Kiszka B., Duda M. Signatures of miR-181a on the renal transcriptome and blood pressure. Mol. Med. 2015;21(1):739–748. doi: 10.2119/molmed.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bozgeyik I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene. 2021;27 doi: 10.1016/j.mgene.2020.100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Trojanowicz B., Imdahl T., Ulrich C., Fiedler R., Girndt M. Circulating miR-421 targeting leucocytic angiotensin converting enzyme 2 is elevated in patients with chronic kidney disease. Nephron. 2019;141(1):61–74. doi: 10.1159/000493805. [DOI] [PubMed] [Google Scholar]
- 133.Zhang C., Wang J., Ma X., Wang W., Zhao B., Chen Y., Chen C., Bihl J.C. ACE2-EPC-EXs protect ageing ECs against hypoxia/reoxygenation-induced injury through the miR-18a/Nox2/ROS pathway. J. Cell Mol. Med. 2018;22(3):1873–1882. doi: 10.1111/jcmm.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct. Target Ther. 2020;5(1):1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Höbel S., Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Internhip Rev. Nanomed. Nanobiotechnol. 2013;5(5):484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 136.Chen Y., He Y., Wang X., Lu F., Gao J. Adipose- derived mesenchymal stem cells exhibit tumor tropism and promote tumorsphere formation of breast cancer cells. Oncol. Rep. 2019;41(4):2126–2136. doi: 10.3892/or.2019.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lotfinegad P. Immunomodulatory nature and site specific affinity of mesenchymal stem cells: a hope in cell therapy. Adv. Pharm. Bull. 2014;4(1):5–13. doi: 10.5681/apb.2014.002. [DOI] [PMC free article] [PubMed] [Google Scholar]