FIG. 2.
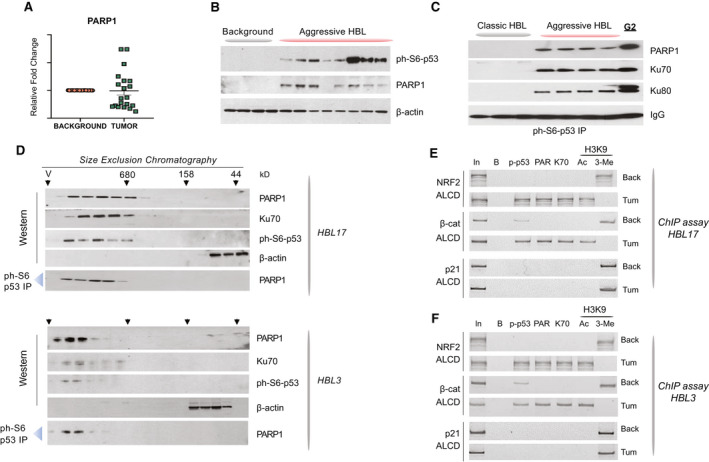
Ph‐S6‐p53/PARP1 complexes are abundant in aggressive HBL. (A) qRT‐PCR‐based analysis of PARP1 in a fresh Bio Bank. (B) PARP1 and ph‐S6‐p53 are elevated in aggressive HBL. WB was performed with nuclear extracts from background and from tumors of aggressive HBL. (C) Co‐IP studies showing that ph‐S6‐p53 forms complexes with PARP1/Ku70 in aggressive HBL and in HepG2 cells. Ph‐S6‐p53 was immunoprecipitated from tumors of mild (classic) HBL and from tumors of aggressive HBL. PARP1, Ku70, and Ku80 were examined in these IPs by WB. (D) HPLC‐based SEC was performed with tumor sections of two aggressive HBL samples. After fractionation, proteins were detected in each fraction by WB. Co‐IP from each fraction shows that ph‐S6‐p53/PARP1 complexes are detected in high MW regions of SEC. (E,F) The ph‐S6‐p53‐PARP1/Ku70 complexes occupy ALCDs of NRF2 and beta‐catenin genes, but not “inactive” ALCD within the p21 gene in 2 additional patients with aggressive HBL. ChIP assay was performed as described in Fig. 1E.
