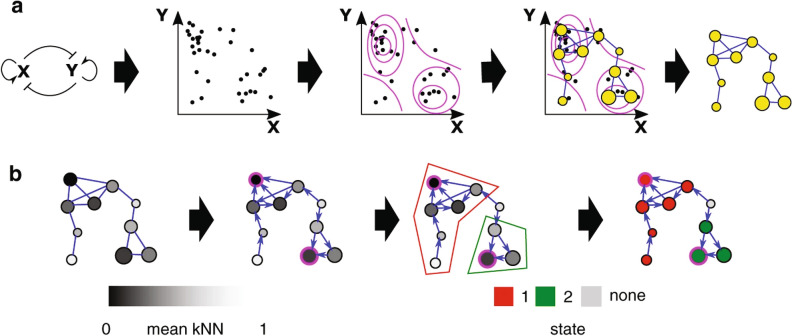
Figure 1.
Microbial communities as metastable dynamical systems. This illustration of the Mapper algorithm is adopted without modifications from25 and is licensed under CC BY 4.0. (a) A community with two species X and Y, that inhibit each other’s growth. The interactions between species make some community compositions more likely than others: either one species is very abundant at the cost of the other, or vice versa. If those configurations are equilibrium states and stable against small perturbations (i.e., if they attract all configurations in their vicinity), they can be considered attractors. The mapper plot can be used to find attractors: It groups samples into clusters (yellow dots) according to their similarity. Clusters with similar compositions are plotted close to each other. The same data-point (a sampled community) can belong to more than one cluster. Then, it connects the clusters that share communities (samples) making a network. Because clusters have different numbers of samples, those with the highest local number of samples potentially are attractors. (b) Finding the attractors in the mapper network. Each node is assigned a k-nearest neighbours (kNN) value (inversely proportional to the density of points) that is later modified to get a potential. The arrows are then directed from higher to lower potential. The nodes with the lowest potentials correspond to attractors, and the nodes that lead to them form their corresponding basin of attraction (all the community states that eventually converge to the attractor). The outcome shows two attractors that correspond to either X or Y with high abundance and the other excluded.