Abstract
Zinc finger E-box-binding homeobox 1 (ZEB1), a functional protein of zinc finger family, was aberrant expressed in many kinds of liver disease including hepatic fibrosis and Hepatitis C virus. Bioinformatics results showed that ZEB1 was abnormally expressed in HCC tissues. However, to date, the potential regulatory role and molecular mechanisms of ZEB1 are still unclear in the occurrence and development of HCC. This study demonstrated that the expression level of ZEB1 was significantly elevated both in liver tissues of HCC patients and cell lines (HepG2 and SMMC-7721 cells). Moreover, ZEB1 could promote the proliferation, migration, and invasion of HCC cells. On the downstream regulation mechanism, ZEB1 could activate the Wnt/β-catenin signaling pathway by upregulating the protein expression levels of β-catenin, c-Myc, and cyclin D1. Novel studies showed that miR-708 particularly targeted ZEB1 3′-UTR regions and inhibited the HCC cell proliferation, migration, and invasion. Furthermore, results of nude mice experiments of HCC model indicated that miR-708 could inhibit tumor growth and xenograft metastasis model was established to validate that miR-708 could inhibit HCC cell metastasis through tail-vein injection in vivo. Together, the study suggested that ZEB1 modulated by miR-708 might be a potential therapeutic target for HCC therapy.
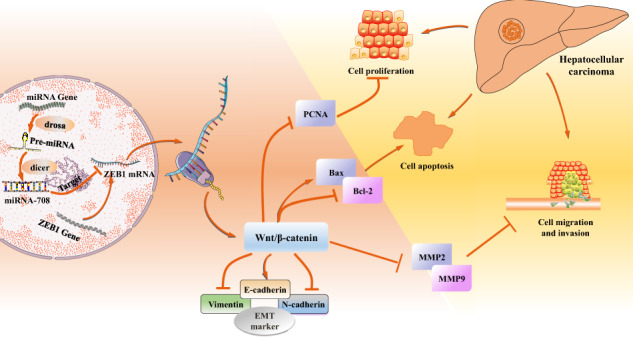
Keywords: ZEB1, HCC, miR-708, Wnt/β-catenin, xenograft metastasis
Introduction
According to the World Health Organization, HCC is the 5th most common cancer and the 2nd most common cause of cancer-related mortality worldwide [1, 2]. HCC has the characteristics of invasion, metastasis, and frequent recurrence, accounting for 90% of primary liver cancer, and has become a major health problem around the world [3]. There are many ways for HCC treatment including medication and targeted therapy, but the survival rate of HCC patients is still very low [4–6]. Therefore, understanding the potential mechanisms of HCC initiation and progression is critical to explore new therapeutic targets for improving the survival rate of HCC patients. The development of HCC is associated with these major factors: viral infections, hereditary disorders, chemical toxins, and metabolic syndromes [7]. These factors regulate the function of oxidative stress, the inflammatory/immune system, and the epithelial–mesenchymal transition (EMT) [8]. EMT is a process in which an epithelial cell transforms into a mesenchymal cell and is involved in wound healing, fibrosis, and metastasis (migration and invasion), leading to tumor progression [9–12]. Furthermore, it is well known that the induction of EMT markers (such as E-cadherin, Vimentin, and N-cadherin) promote the development and progression of cancer [13, 14].
As an activator of EMT, ZEB1, the member of zinc finger family, encodes the zinc finger and homeodomain transcription cytokine [15–17]. Numerous studies have demonstrated that abnormal expression of ZEB1 in many types of liver disease including hepatitis, liver fibrosis, and hepatitis C virus [18–20]. For example, ZEB1 was upregulated in liver fibrosis and increased the secretion of IL-6 and TNF-α via the Wnt/β-catenin signaling pathway [19]. The process of these diseases will develop into HCC if the damage factor cannot be removed for a long time [21]. Thereby, it is valuable to investigate the function of ZEB1 in HCC. HCC is one of the digestive system cancers, in other digestive system cancers, recent studies indicated that ZEB1 increased the invasive activity of gallbladder cancer cell [22]. In human colon cancer cell lines, ZEB1 promoted the ability of invasive and migratory in vitro and metastases in vivo. In addition, previous studies demonstrated that ZEB1 activated EMT and played a key driving force in the development of pancreatic tumors from early tumorigenesis to late metastasis [23, 24]. Increased evidence has shown that ZEB1 plays a regulator role in metastasis of digestive system cancer. Based on these observations, we hypothesized that ZEB1 may serve a significant role in the tumorigenesis of HCC by activating EMT [25–27].
MicroRNAs (miRNAs), short 20–22 nucleotides, were recognized to hold essential parts in tumor progression by binding to the 3′-untranslated region (3′-UTR) of the target mRNAs [28–31]. Analysis of miRNA expression has identified a group of dysregulated miRNAs in HCC [32]. According to the bioinformatics results (TargetScan and miRDB) and our previous study showed that ZEB1 may be a target of miR-708. MiR-708 could specifically interact with the 3′-UTR of ZEB1 mRNA to inhibit the expression of ZEB1 [33]. Therefore, the study explored the function role of ZEB1 in HCC progression and regulatory mechanism. This study found that ZEB1 dramatically enhanced the Wnt/β-catenin signaling pathway and thereby aggravated HCC, which was regulated by miR-708.
Materials and methods
Specimen collection
The liver tissues were obtained from HCC patients undergoing partial liver resection from the First Affiliated Hospital of Anhui Medical University. Hematoxylin and eosin (H&E) was used to detect the pathology of HCC tissues. The characteristics of the patients are shown in Table 1. Clinically, we collected 370 HCC patients with different stages and analyzed the expression level of ZEB1, as shown in Table 2. The study was complied with the standards set by the Helsinki Declaration approved by the Ethics Committee of Health Medical Research (20190246) of Anhui Medical University. All participants signed patients’ informed consent.
Table 1.
Characteristics of the analyzed HCC patients.
| Characteristics | Category | Number | % |
|---|---|---|---|
| Gender | Female | 24 | 60 |
| Male | 16 | 40 | |
| Ages (year) | 30–50 | 4 | 10 |
| 50–70 | 22 | 55 | |
| >70 | 14 | 35 | |
| T stage | T1 | 1 | 2.5 |
| T2 | 8 | 20 | |
| T3 | 15 | 37.5 | |
| T4 | 6 | 15 | |
| Unknown | 10 | 25 | |
| AFP (ng/mL) | 0–7 | 12 | 30 |
| >7 | 28 | 70 | |
| CEA (ng/mL) | 0–6.5 | 25 | 62.5 |
| >6.5 | 15 | 37.5 | |
| LYMPH (%) | <20 | 31 | 77.5 |
| 20–50 | 9 | 22.5 | |
| HCT (%) | <40 | 22 | 55 |
| 40–50 | 18 | 45 | |
| MCV (g) | 80–100 | 30 | 75 |
| >100 | 10 | 25 |
Table 2.
Association of ZEB1 expression with the clinicopathological parameters from HCC patients.
| Variables | Low ZEB1 group n = 162 |
High ZEB1 group n = 208 |
χ2 | P value |
|---|---|---|---|---|
| Age(year) | 0.538 | 0.463 | ||
| ≤ 60 | 74 | 103 | ||
| >60 | 88 | 105 | ||
| Gender | 1.237 | 0.266 | ||
| Male | 114 | 135 | ||
| Female | 48 | 73 | ||
| Metastasis | 1.890 | 0.389 | ||
| M0 | 122 | 144 | ||
| M1 | 2 | 2 | ||
| Mx | 38 | 62 | ||
| AJCC stage | 5.495 | 0.064 | ||
| I/II | 122 | 134 | ||
| III/IV | 33 | 57 | ||
| Null | 7 | 17 | ||
| Histologic grade | 4.146 | 0.126 | ||
| G1/G2 | 105 | 127 | ||
| G3/G4 | 57 | 76 | ||
| Null | 0 | 5 | ||
| Survival status | 6.845 | 0.009 | ||
| Alive | 117 | 123 | ||
| Dead | 45 | 85 |
Bold values indicate statistical significance. If the P value <0.05, the data were considered significant difference, and if the P value <0.01, the data were considered different with strong significance.
Magnetic resonance imaging (MRI)
The patients received MRI using a 1.5-T (Magnetom Avanto, Siemens Healthcare). For enhanced MRI scanning, patients were injected with 0.025-mmol/kg gadoxetate disodium (Primovist; Bayer HealthCare), followed by saline irrigation. T1-weighted three-dimensional gradient echo imaging was acquired before contrast injection and in the arterial, portal (50 s), transitional (3 min), and hepatobiliary phases (20 min) of contrast enhancement.
Cell culture
SMMC-7721 and HepG2 cells were purchased from Cell Bank at the Chinese Academy of Sciences (Shanghai, China). Cells were cultured according to previous study [34]. When the growth rate of monolayer cells reached 80%, 0.25% of trypsin-treated cells was added to divide SMMC-7721 cells into 2 groups, namely, high expression miR-708 group (LV-miR-708 group) and negative control group (LV-miR-NC group). Appropriate amount of lentivirus is added in the logarithmic growth phase of liver cancer cells. After 3 days of infection, the expression of the reporter gene GFP mediated by the lentiviral vector was observed. When the infection rate reached above 95%, cells were further cultured for subsequent experiments. hsa-miR-708 (MI0005543) lentiviral vector was purchased from HanBio Technology (Shanghai, China).
Immunohistochemistry (IHC)
Ten percent neutral buffered formalin solution was used to fixate HCC tissues for 24 h, after embedding in paraffin, and stained for routine histology. HCC sections were deparaffinized with xylene and dehydrated in decreasing concentrations of ethanol. Microwaving in citric saline for 15 min was used for antigen recovery. The tissues were then blocked with 2% bovine serum albumin (BSA) by incubation with primary antibodies against ZEB1 (1:100, Abcam, UK) for 12 h at 4 °C. Then the HCC tissues were incubated with the secondary antibody at 37 °C for 60 min. The expression level of ZEB1 in the tissues was observed by 3,3′-diaminobenzidine tetrahydrochloride staining. The sections were subsequently counterstained with hematoxylin for 5 min. After fixed with a gel, the microscope was used for imaging.
Luciferase reporter assay
pHG-MirTarget-ZEB1-3′UTR plasmid and mutant 3′UTR-ZEB1 were obtained from HonorGene Biotechnology (Changsha, Hunan, China). HepG2 and SMMC-7721 cells were inoculated in a six-well plate in advance. After that, ZEB1-3′UTR-WT plasmid gene (200 ng), miR-708 mimics, or NC control (60 nmol) were chosen, the same experiment was performed in the control group of mutant 3′UTR-ZEB1. The luciferase activity was detected by using the luciferase assay kit (Promega Corporation, Madison, USA).
RNA interference analysis
HepG2 and SMMC-7721 cells were seeded in six-well plate before transfecting with ZEB1 siRNA (GenePharma, Shanghai, China) and Negative Control siRNA (NC) (GenePharma, Shanghai, China) by using LipofectamineTM2000 (Invitrogen, California, USA) according to the manufacturer’s protocol. The sequences are as following:
ZEB1 siRNA: F: 5′-GUCGCUACAAACAGUUGUATT-3′, R: 5′-UACAACUGUUUGUAGCGACTT-3′.
NC siRNA: F: 5′-UUCUCCGAACGUGUCACGUTT-3′, R: 5′-ACGUGACACGUUCGGAGAATT-3′.
miR-708 mimics: F: 5′-AAGGAGCUUACAAUCUAGCUGGG-3′, R: 5′-CAGCUAGAUUGUAAGCUCCUUUU-3′.
miR-708 mimics NC: F: 5′-UUCUCCGAACGUGUCACGUTT-3′, R: 5′-ACGUGACACGUUCGGAGAATT-3′.
miR-708 inhibitor: 5′-CCCAGCUAGAUUGUAAGCUCCUU-3′.
miR-708 inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′.
Plasmid transfection
Overexpression plasmid of ZEB1 (P-CMV-ZEB1) was constructed and stored in our laboratory (Anhui, China) [19]. The P-CMV vector was used as an internal control. P-CMV-ZEB1 and P-CMV were transfected in HepG2 and SMMC-7721 cells by using Lipofectamine™2000 in accordance with the manufacturer’s instructions.
Western blotting
HepG2 and SMMC-7721 cells (1 × 105) were seeded in the six-well plate and washed three times via cold phosphate-buffered saline (PBS). Cells underwent cytolysis to total protein by protein lysate (RIPA lysate: PMSF = 100:1) on ice for 30 min. The protein was extracted and subsequently was electrophoresed for 1 h. After electrophoresis, the protein was blotted onto the PVDF membrane by electrotransfer. After blocking the nonspecific protein binding by BSA, the PVDF membrane was incubated overnight in the diluted primary antibody, and the primary antibody dilution ratios were ZEB1 (1:1000), Bax (1:1000), Bcl-2 (1:1000), PCNA (1:1000), MMP2 (1:1000), MMP9 (1:1000), E-cadherin (1:1000), N-cadherin (1:1000), Vimentin (1:1000), respectively. The protein was incubated for 1 h with the secondary antibody, and then the immune-reactive bands were detected by using the ECL-chemiluminescent kit (ECL-plus, Thermo Fisher Scientific) [35].
Transwell migration assay
The bottom chamber of the Transwell system (Corning, NY, USA) is filled with 500 μL of 20% FBS of culture medium. The cell suspension in serum-free medium (200 μL, 4 × 104 cells) was added to the upper chamber. The cells were fixed with 10% neutral buffered formalin solution and colored with 0.1% crystal violet after incubation for 24 h. The number of cells penetrating the membrane was counted in five random fields. All experiments were repeated three times.
EdU DNA incorporation assay
Cell proliferation was detected by using standard EdU DNA incorporation assay. Exponentially growing HepG2 and SMMC-7721 cells were plated on 13-mm glass coverslips, respectively. Then, P-CMV-ZEB1, ZEB1-siRNA, miR-708 mimics, miR-708 inhibitor and there NC were transfected into HepG2 and SMMC-7721 cells with LipofectamineTM2000, respectively. After culture 24 h, the cells were labeled with 50-μm EdU for 2 h, and then rinsed twice with ice-cold PBS. After labeling, HepG2 and SMMC-7721 cells were fixed in 4% paraformaldehyde for 30 min and rinsed with 2-mg/mL glycine for 5 min, respectively, then permeabilised with 1% Triton X-100 for 10 min, rinsed with PBS for 5 min. Apollo staining was performed for 30 min and then Hoechst staining for 30 min. The images were taken by fluorescence microscopy (Olympus, Tokyo, Japan).
Transwell invasion assay
HepG2 and SMMC-7721 cells were inoculated into the upper chamber with a serum-free medium at a density of 2 × 105 cells/well. The bottom chamber is filled with 500 µL of 20% FBS culture medium. After incubated in a 5% (v/v) CO2 incubator at room temperature for 2 days, the noninvasive cells and matrigel were removed in the upper chamber, and then the cells were fixed on the lower surface with 10% neutral buffered formalin solution and stained with 0.1% crystal violet . The invading cells was counted in five randomly selected microscope fields. The experiments were repeated three times.
Cell-cycle analysis
HepG2 and SMMC-7721 cells were, respectively, transfected with P-CMV-ZEB1 and ZEB1 siRNA and their NC for 48 h. Before lysed, the cells were cleaned with PBS and fixed with 75% cold ethanol for 12 h at 4 °C. After centrifugation (1000 ×g, 5 min) and resuspension, the cells were incubated with RNase and propidium iodide (PI) (Beyotime, Shanghai, China) at room temperature for 30 min protecting from light. The result was analyzed by the flow assay (BD Biosciences, NJ, USA) and ModFit software (Verity Software House, USA).
Flow cytometry
HepG2 and SMMC-7721 cells at logarithmic growth phase (2 × 105/mL) were seeded in a six-well plate until grew with adherence. The cells were digested by trypsin and centrifuged at 1000 rpm for 5 min. After wash with cold PBS, the cells were resuspended in 1× binding buffer, then stained with 5-µL FITC Annexin V and 5-µL PI. The flow cytometer (Beckman Coulter, USA) was used to detect the apoptosis.
Xenograft tumor model in nude mice
Six-week-old male BALB/c nude mice were purchased and reared under the Experimental Animal Center of Anhui Medical University (Hefei, China). The animal testing procedures have been approved by the Code of Ethics and reviewed and implemented according to the guidelines of the Animal Care and Use Committee of Anhui Medical University (number: LLSC20150348). The nude mice were randomly divided into two groups: LV-miR-708 and LV-miR-NC. After that, 200 µL of SMMC-7721 (LV-miR-708/ LV-miR-NC) cells (5 × 106) were subcutaneously injected into the right flank of each mouse. Eight weeks later, the mice were sacrificed and the tumors were dissected for further evaluation.
Metastasis model
Thirty-six 5-week-old nude mice were housed under specific pathogen-free conditions. SMMC-7721 cells were infected with lentivirus miR-708-Null-LUC-PURO and LUC-PURO (Hanbio Biotechnology, Shanghai, China), respectively, and puromycin was used to screen out the cells infected with lentivirus successfully. SMMC-7721/miR-708-Null-LUC-PURO and SMMC-7721/LUC-PURO cells (1 × 106) were injected into the tail veins of mice to evaluate the effect of miR-708 on the metastasis ability of HCC cells. After an intraperitoneal injection of luciferin (P1043, Promega, USA) (150 mg/kg), the mice were anaesthetized with 10% chloral hydrate. The bioluminescence of metastatic tumors was detected within 15 min. An IVIS Lumina III Imaging System (Caliper Life Sciences, USA) was used to observe the quantity and location of metastatic tumors in living mice at 30 min, 1 week, and 3 weeks after the tumor cells were injected. The animal testing procedures have been approved by the Code of Ethics and reviewed and implemented according to the guidelines of the Animal Care and Use Committee of Anhui Medical University (number: LLSC20150348).
Statistical analysis
Statistical data analysis was used by SPSS ver.18.0. The differences in groups were checked by one-way ANOVA. The data were performed as the mean ± standard error at least three times independently. If the P value < 0.05, the data were considered significantly different, and if the P value < 0.01, the data were considered with strongly significant difference.
Results
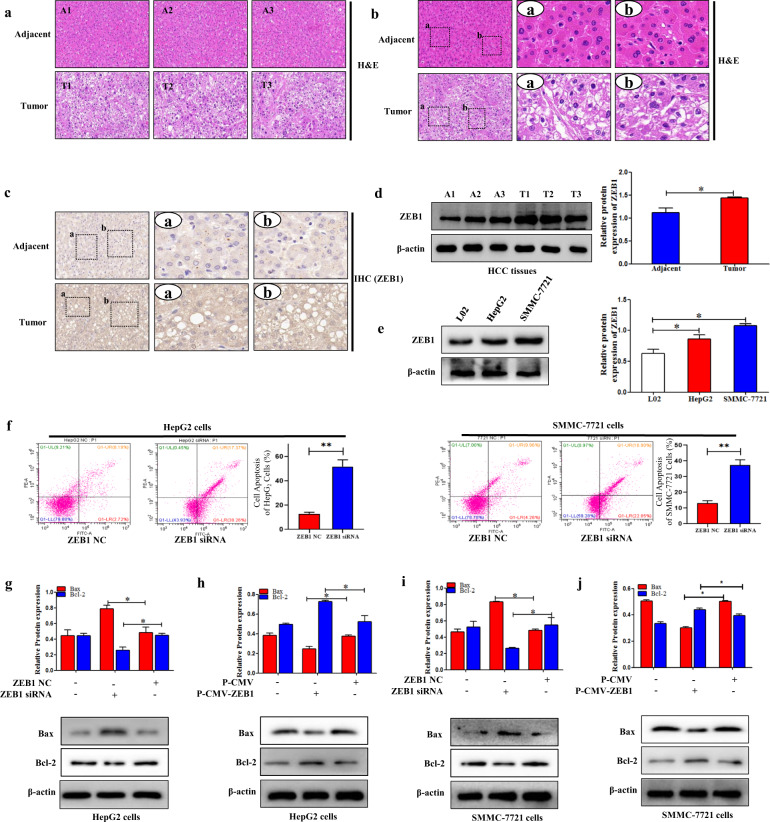
ZEB1 was aberrantly expressed in human HCC tissues
To investigate the role of ZEB1 in human HCC, the liver tissues of HCC patients were obtained for the study. First, the results of MRI showed that the size and shape of liver were irregular and the liver fissures were widened. There were multiple low-density nodules and masses scattered in the liver parenchyma. The portal vein wall was slightly rough (Fig. S1a). Second, the results of H&E staining displayed that there was a large area of necrosis in HCC tissues (Fig. 1a, b). In addition, the results of TCGA showed that the mRNA expression of ZEB1 was higher in HCC tissues compared to the normal tissues, and the overall survival was significantly reduced compared to the healthy people (Fig. S1b,c), suggesting that ZEB1 might play a key role in the progression of HCC. More importantly, the cutoff value is determined by the ROC curve to ensure that the Yoden Index is maximized. Patients in the high-risk group had a significantly worse prognosis than those in the low-risk group (Fig. S1d and Table 1). Clinically, we collected 370 HCC patients with different stages and analyzed the expression level of ZEB1. Statistical analysis showed that higher expression of ZEB1 in HCC tissue was positively associated with survival status (P = 0.009). However, there was no significant relationship between the degrees of ZEB1 staining and metastasis, AJCC stage, histologic grade, age, or the number of tumor (Fig. S1e and Table 2). In general, the results suggested that ZEB1 was aberrantly expressed in human HCC tissues.
Fig. 1. ZEB1 was aberrantly expressed in human HCC tissues.
a, b The H&E stain in human HCC tissues and adjacent tissues. The rectangular image in the left panel was magnified in the middle and right panels. The scale was shown in the figure. Pannoramic SCAN 150 (3DHISTECH, Budapest, Hungary) was used for the imaging. c The IHC of ZEB1 in human HCC tissues and adjacent tissues. The rectangular image in the left panel was magnified in the middle and right panels. The scale was shown in the figure. Pannoramic SCAN 150 (3DHISTECH, Budapest, Hungary) was used for the imaging. d The protein expression level of ZEB1 was measured by using Western blotting in human HCC tissues compared to the adjacent tissues. e The protein expression level of ZEB1 was measured by using Western blotting in L02, HepG2, SMMC-7721 cells. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group. ZEB1 inhibited the apoptosis of HCC cells. f Cell apoptosis was measured by using flow cytometry analysis in HepG2 and SMMC-7721 cells transfected with ZEB1 siRNA. g–j The protein expression levels of Bax, Bcl-2, were measured by using Western blotting analysis in HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
ZEB1 was increased in HCC tissues and HCC cell lines
In the study, the expression level of ZEB1 was first analyzed in HCC tissues and adjacent tissues. Results of IHC indicated that the expression level of ZEB1 was upregulated in HCC tissues compared to the adjacent tissues (Fig. 1c). These results were confirmed by using Western blotting analysis (Fig. 1d). Then, the protein expression level of ZEB1 was detected in HepG2, SMMC-7721, and L02 cells. The results showed that the expression level of ZEB1 in HCC cell lines was significantly increased compared to L02 cells (Fig. 1e). In general, the finding revealed that ZEB1 was increased in HCC tissues and HCC cell lines.
ZEB1 inhibited apoptosis of HCC cells in vitro
To detect the role of ZEB1 on cell apoptosis, ZEB1-siRNA and P-CMV-ZEB1 were transfected, respectively, in HepG2 and SMMC-7721 cells to decrease and increase the expression level of ZEB1. The flow cytometry results showed that ZEB1-siRNA significantly promoted cell apoptosis both in HepG2 and SMMC-7721 cells (Fig. 1f). Moreover, Western blotting results showed that ZEB1-siRNA increased the expression level of Bax and inhibited the expression level of Bcl-2 in HepG2 and SMMC-7721 cells (Fig. 1g, i). Interestingly, overexpression of ZEB1 remarkably inhibited the expression level of Bax and promoted the expression level of Bcl-2 (Fig. 1h, j). Collectively, these results provided evidence that ZEB1 could inhibit cells apoptosis in HepG2 and SMMC-7721 cells.
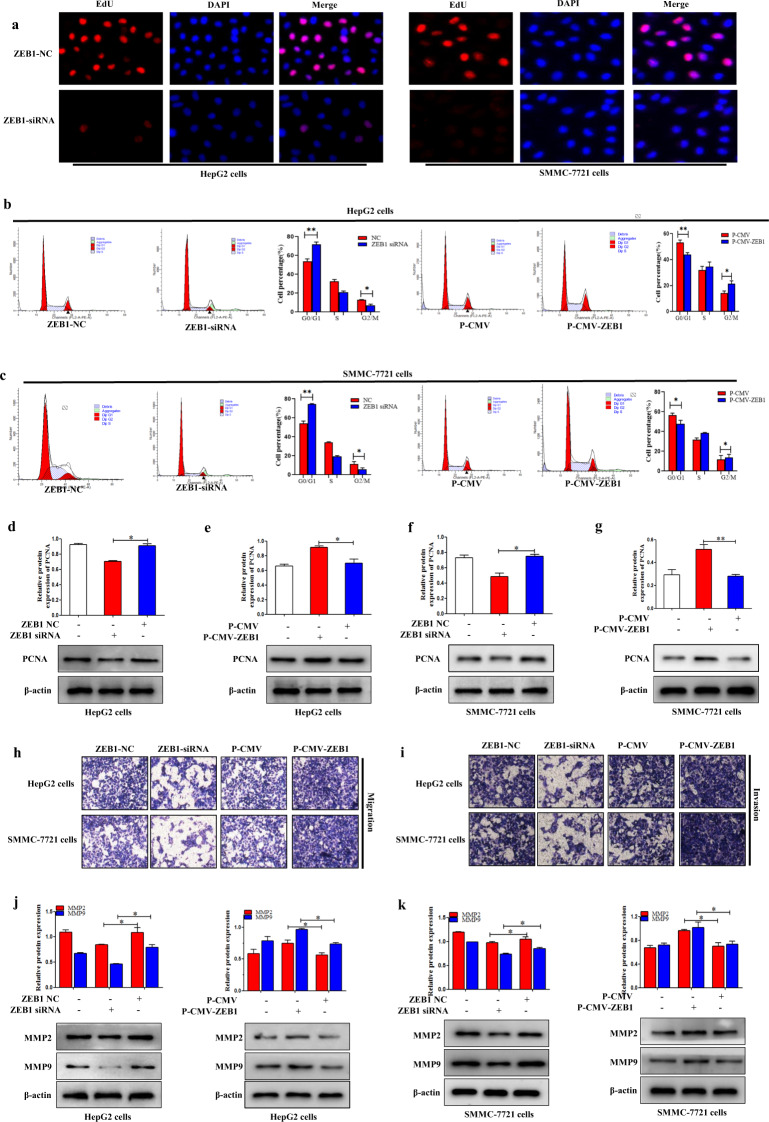
ZEB1 promoted proliferation of HCC cells
EdU staining was used to detect the function of ZEB1 on the proliferation of HepG2 and SMMC-7721 cells. The results showed that ZEB1 siRNA significantly reduced the proliferation of HepG2 and SMMC-7721 cells compared to the ZEB1 NC group (Fig. 2a). In addition, the study found that ZEB1 siRNA could obviously reduce the expression level of cell proliferation marker PCNA (Fig. 2d,f). In contrast, P-CMV-ZEB1 in HepG2 and SMMC-7721 cells could lead to higher protein expression level of PCNA compared to the control group (Fig. 2e, g). Next, the function of ZEB1 on the HCC cell proliferation was detected by using the cell-cycle assay. The flow cytometry results showed that ZEB1 siRNA significantly increased the number of cells in the G0/G1 phase but decreased the number in the G2/M phase of HepG2 and SMMC-7721 cells compared to the NC group (Fig. 2b, c), indicating that ZEB1 siRNA inhibited cell-cycle progression. In contrast, the percentage of cells in the G2/M phase of HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 was increased compared to the control group (Fig. 2b, c). In summary, these results indicated that ZEB1 can actively promote the proliferation of HCC cells.
Fig. 2. ZEB1 promoted the proliferation of HCC cells.
a Cell proliferation was measured by using the EdU assay in HepG2 and SMMC-7721 cells transfected with ZEB1. Fluorescence microscope was used for the imaging. b, c Cell arrest in G0/G1 phase in HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. d–g The protein expression level of PCNA was measured by using Western blotting analysis in HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group. ZEB1 promoted the migration and invasion of HCC cells. h Transwell migration assay of migratory capacity of HepG2 and SMMC-7721 cells was transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. i Matrigel Transwell assay of invasive potential of HepG2 and SMMC-7721 cells was transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. j, k The protein expression levels of MMP2 and MMP9 were measured by using Western blotting in HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
ZEB1 promoted migration and invasion of HCC cells
A series of detection methods are used to detect the effect of ZEB1 on the migration and invasion of HCC cells. Transwell migration analysis showed that ZEB1 siRNA caused significantly less migration than the control group in HepG2 and SMMC-7721 cells (Fig. 2h). Next, the cells invasion was detected by using the Matrigel-coated Transwell invasion test. The results showed that ZEB1 siRNA reduced the ability of cells to invade through the gel matrix into the adjacent chamber compared to the control group (Fig. 2i). Conversely, P-CMV-ZEB1 increased cell migration and invasion in HepG2 and SMMC-7721 cells (Fig. 2h, i). Moreover, the results showed that P-CMV-ZEB1 significantly increased the protein expression levels of MMP2/MMP9 in HepG2 and SMMC-7721 cells (Fig. 2j, k), and the function of P-CMV-ZEB1 on MMP2/MMP9 was reversed by knocking down ZEB1 (Fig. 2j, k). Taken together, these data revealed that ZEB1 potentially promotes the migration and invasion of HCC in vitro.
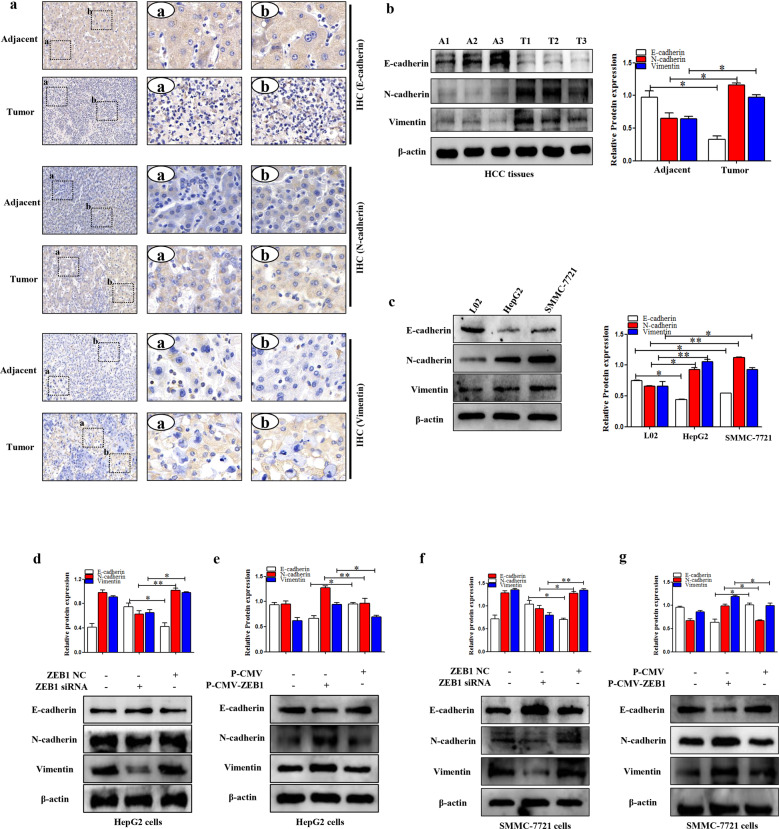
ZEB1 aggravated the process of EMT in HCC
EMT is essential for tumor-cell survival [36]. Thus, the relative proteins of EMT were detected both in human HCC tissues and HCC cell lines (HepG2 and SMMC-7721 cells). First, IHC and Western blotting results indicated that the expression levels of N-cadherin and Vimentin were upregulated whereas the expression level of E-cadherin was downregulated both in human HCC tissues and HCC cell lines (HepG2 and SMMC-7721 cells) (Fig. 3a–c). Subsequently, the function of the ZEB1 on EMT was detected in HepG2 and SMMC-7721 cells. ZEB1 siRNA promoted the expression level of E-cadherin whereas inhibited the expression levels of N-cadherin and Vimentin (Fig. 3d,f). Notably, the function of ZEB1 siRNA on EMT could be reversed by P-CMV-ZEB1 (Fig. 3e,g). Collectively, the data indicated that ZEB1 could aggravate the process of EMT both in human HCC tissues and cell lines (HepG2 and SMMC-7721 cells).
Fig. 3. ZEB1 promoted the process of EMT in HCC.
a IHC of E-cadherin, N-cadherin, and Vimentin in human HCC tissues and adjacent tissues. b Western blotting analysis of E-cadherin, N-cadherin, and Vimentin in human HCC tissues and adjacent tissues. c Western blotting analysis of E-cadherin, N-cadherin, and Vimentin in L02, HepG2, and SMMC-7721 cells. d–g The protein expression levels of E-cadherin, N-cadherin, and Vimentin were detected in HepG2 and SMMC-7721 cells transfected with P-CMV-ZEB1 and ZEB1 siRNA, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
ZEB1 is a direct target of miR-708 in HCC cells
Bioinformatics tools (TargetScan and miRDB) were used to predict the target genes of miR-708. The miRNA target prediction algorithm predicted that the 3′-UTR of ZEB1 mRNA contains putative miR-708 binding sites (Fig. S2a). The 3′-UTR of ZEB1 was cloned from it into the pmirGLO luciferase reporter vector to confirm that whether miR-708 could regulate ZEB1 by combining to the corresponding 3′-UTR. Then, the miR-708 or control mimics were co-transfected with P-CMV-ZEB1 into HepG2 cells. Double luciferase reporter assay was subsequently used to evaluate ZEB1 response to miR-708 (Fig. S2b). These findings revealed that miR-708 mimics obviously declined ZEB1 luciferase activity in HepG2 cells. Moreover, Western blotting result demonstrated that miR-708 mimics downregulated the expression level of ZEB1 in HepG2 cells. Conversely, miR-708 inhibitor upregulated the expression level of ZEB1 in HepG2 cells (Fig. S2c). Next, miR-708 mimics and P-CMV-ZEB1 were co-transfected in HepG2 cells. Western blotting result revealed that the protein levels of Bcl-2, MMP2/MMP9, and PCNA were obviously decreased in the co-transfection group compared to the group transfected with P-CMV-ZEB1. Meanwhile, the expression level of Bax in the co-transfection group was significantly higher than that in the group transfected with P-CMV-ZEB1 (Fig. S2d). Thus, these results further indicated that ZEB1 is a direct target of miR-708 in HepG2 cells.
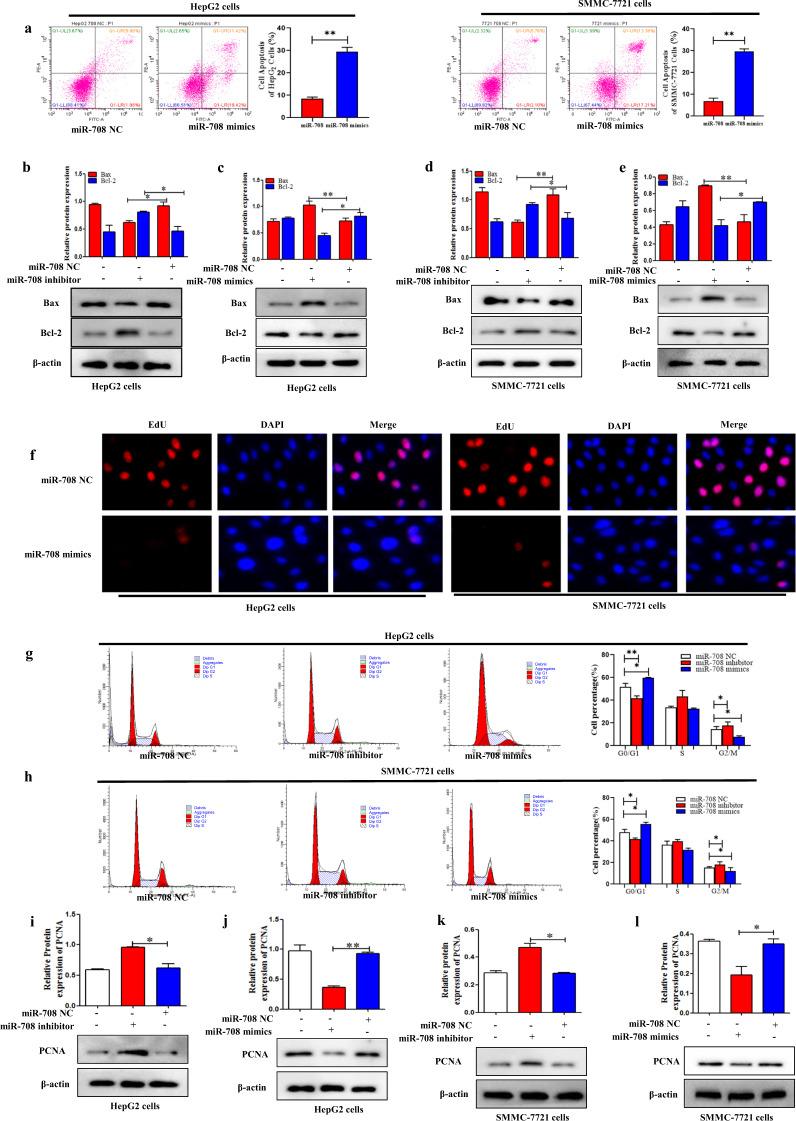
MiR-708 promoted apoptosis of HCC cells
To confirm the function of miR-708 on the cell apoptosis in HCC cells, RT-qPCR result indicated that the expression level of miR-708 was downregulated both in human HCC tissues and HepG2 and SMMC-7721 cells compared to the adjacent tissues and L02 cells, respectively (Fig. S2e, f). After the establishment of miR-708 overexpression HCC line, the flow cytometry result indicated that miR-708 mimics significantly promoted cell apoptosis both in HepG2 and SMMC-7721 cells (Fig. 4a). Moreover, the results showed that miR-708 mimics significantly inhibited the expression level of Bcl-2, whereas promoted the expression level of Bax in HepG2 and SMMC-7721 cells (Fig. 4c, e). Importantly, the function of miR-708 mimics on cell apoptosis was reversed by miR-708 inhibitor (Fig. 4b, d). Collectively, these results provided evidence that miR-708 could promote cell apoptosis in HepG2 and SMMC-7721 cells.
Fig. 4. MiR-708 promoted the apoptosis of HCC cells.
a Cell apoptosis was measured by using flow cytometry analysis in HepG2 and SMMC-7721 cells transfected with miR-708 mimics. b–e The protein expression levels of Bax, Bcl-2, were measured by using Western blotting analysis in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group. MiR-708 inhibited the proliferation of HCC cells. f Cell proliferation was determined by using the EdU assay in HepG2 and SMMC-7721 cells transfected with miR-708 mimics. Fluorescence microscope was used for the imaging. g, h Cell arrest in G0/G1 phase in HepG2 and SMMC-7721 cells was detected by flow cytometric analysis transfected with miR-708 mimics and miR-708 inhibitor, respectively. i–l The protein expression level of PCNA was measured by using Western blotting analysis in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
MiR-708 inhibited proliferation of HCC cells
To detect whether miR-708 could affect cell proliferation in HCC cells, EdU staining was used to detect the effect of miR-708 on the proliferation of HepG2 and SMMC-7721 cells. The results showed that miR-708 mimics significantly reduced cell proliferation in HepG2 and SMMC-7721 cells (Fig. 4f). In addition, we found that miR-708 mimics obviously decreased the expression level of PCNA, a marker of cell proliferation (Fig. 4j,l). In contrast, miR-708 inhibitor upregulated the expression level of PCNA compared to the control group in HepG2 and SMMC-7721 cells (Fig. 4i, k). Moreover, the function of miR-708 on HCC cell proliferation was detected by using cell-cycle assay. Flow cytometry assay revealed that miR-708 mimics could increase the larger G0/G1 population and decrease the percentage of G2/M compared to the miR-708 NC group (Fig. 4g,h). In contrast, miR-708 inhibitor could decrease the percentage of cells in the G0/G1 phase and increase the larger G2/M population compared to the control group in HepG2 and SMMC-7721 cells (Fig. 4g, h). These findings demonstrated that miR-708 is negatively correlated with HCC cell proliferation.
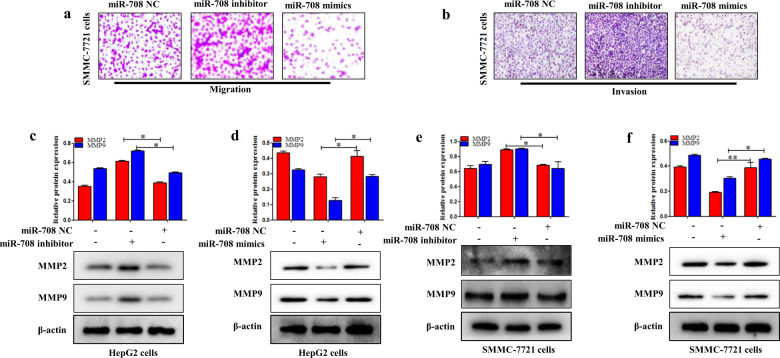
MiR-708 inhibited migration and invasion of HCC cells
Transwell migration and Western blotting analysis were used to detect whether miR-708 affects HCC cell migration and invasion. The results showed that miR-708 mimics significantly caused less migration compared to the control group, and miR-708 inhibitor caused significantly higher migration compared to the control group (Fig. 5a). Then, Matrigel-coated Transwell invasion test demonstrated that miR-708 mimics decreased the ability of cells to invade across the gel matrix into adjacent chambers compared to the control group (Fig. 5a). In contrast, the miR-708 inhibitor increased cells migration and invasion compared to the control group in SMMC-7721 cells (Fig. 5a,b). Increasingly, evidence demonstrated that TGF-β1 significantly induced EMT to promote migration and invasion of HepG2 cells [37]. HepG2 cells were treated with TGF-β1 5 ng/mL for 24 h [38]. The results showed that TGF-β1 significantly induced EMT to promote migration and invasion of HepG2 cells (Fig. S3a). miR-708 mimics significantly decreased the ability of cells to migrate across the gel matrix into adjacent chambers compared to the miR-708 NC group. In the same way, miR-708 mimics also downregulated HepG2 cell migration and invasion after treated with TGF-β1 5 ng/mL for 24 h, compared to the miR-708 NC group treated with 5 ng/mL TGF-β1 (Fig. S3b). Subsequently, the expression level of MMP2/MMP9 protein was examined in HepG2 and SMMC-7721 cells. The results showed that miR-708 mimics suppressed the expression level of MMP2/MMP9 protein in HepG2 and SMMC-7721 cells (Fig. 5d,f), while miR-708 inhibitor upregulated the expression levels of MMP2/MMP9 (Fig. 5c,e). In summary, miR-708 may inhibit the migration and invasion of HCC cells in vitro.
Fig. 5. MiR-708 inhibited the migration and invasion of HCC cells.
a The migratory capacity was detected by using the Transwell migration assay in SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. b The invasive potential was detected by the Matrigel Transwell assay in SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. c, d The protein expression levels of MMP2 and MMP9 were measured by using Western blotting in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
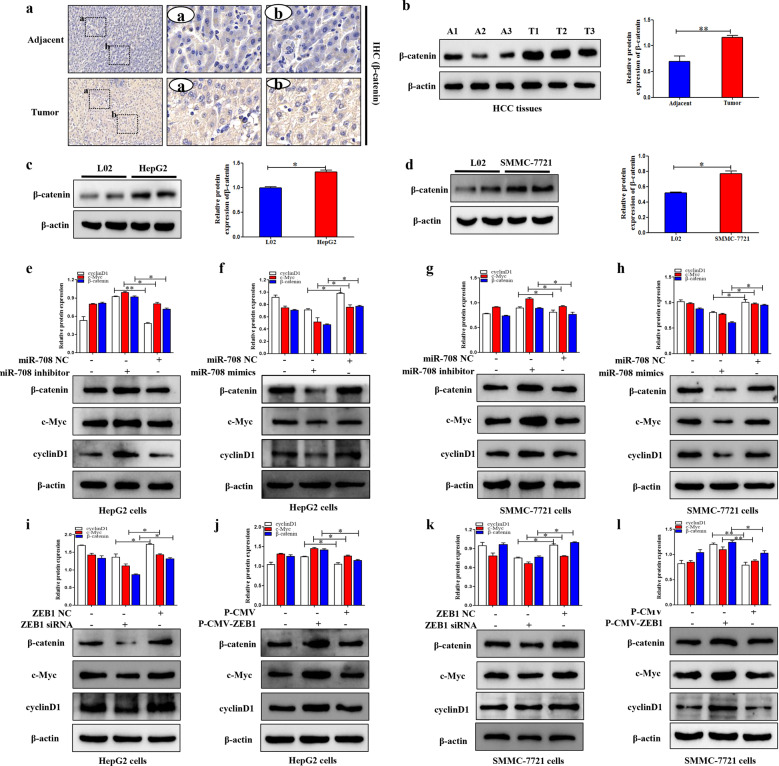
ZEB1-activated Wnt/β-catenin signaling pathway regulated by miR-708 in HCC cells
The activation of Wnt/β-catenin signaling pathway is closely related to the proliferation, invasion, and migration of the progression of tumor. Mechanically, the target genes of the Wnt/β-catenin signaling pathway were examined by using the Western blotting assay. In this study, IHC and Western blotting results showed that the expression level of β-catenin in HCC tissues was higher compared to the control group (Fig. 6a,b). Then, Western blotting result revealed that the protein level of β-catenin in HepG2 and SMMC-7721 cells was higher than that in L02 cells (Fig. 6c,d). In order to detect whether the Wnt/β-catenin signaling pathway was involved in the progression of HCC, the protein levels of β-catenin, and its downstream targets c-Myc and Cyclin D1 were detected in HepG2 and SMMC-7721 cells transfected with ZEB1 siRNA and P-CMV-ZEB1, respectively. ZEB1 siRNA obviously reduced the expression levels of β-catenin, c-Myc, and Cyclin D1 compared to the control group (Fig. 6i,k). Conversely, the expression levels of β-catenin, c-Myc, and cyclin D1 were significantly upregulated in HepG2 and SMMC-7721 cells transfected with p-CMV-ZEB1 compared to the control group (Fig. 6j,l). Subsequently, the expression levels of β-catenin and its downstream targets c-Myc and cyclin D1 were measured in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. The results showed that the miR-708 mimics obviously reduced the expression levels of β-catenin, c-Myc, and cyclin D1 (Fig. 6f,h). The function of miR-708 mimics on the Wnt/β-catenin signaling pathway was reversed by knocking down miR-708 (Fig. 6e,g). In summary, these findings indicated that ZEB1 could positively activate the Wnt/β-catenin signaling pathway, which was regulated by miR-708.
Fig. 6. ZEB1 activated Wnt/β-catenin signaling pathway in HCC cells.
a The IHC of β-catenin in human HCC tissues and adjacent tissues. The rectangular image in the left panel was magnified in the middle and right panels. b The protein expression level of β-catenin in human HCC tissues and adjacent tissues was analyzed by Western blotting. c The protein expression level of β-catenin in L02 and HepG2 cells was analyzed by Western blotting. d The protein expression level of β-catenin in L02 and SMMC-7721 cells was analyzed by Western blotting. e–h The protein expression levels of β-catenin, c-Myc, and cyclinD1 were performed in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. i–l The protein expression levels of β-catenin, c-Myc, and cyclinD1 were detected in HepG2 and SMMC-7721 cells transfected with ZEB1 siRNA and P-CMV-ZEB1, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
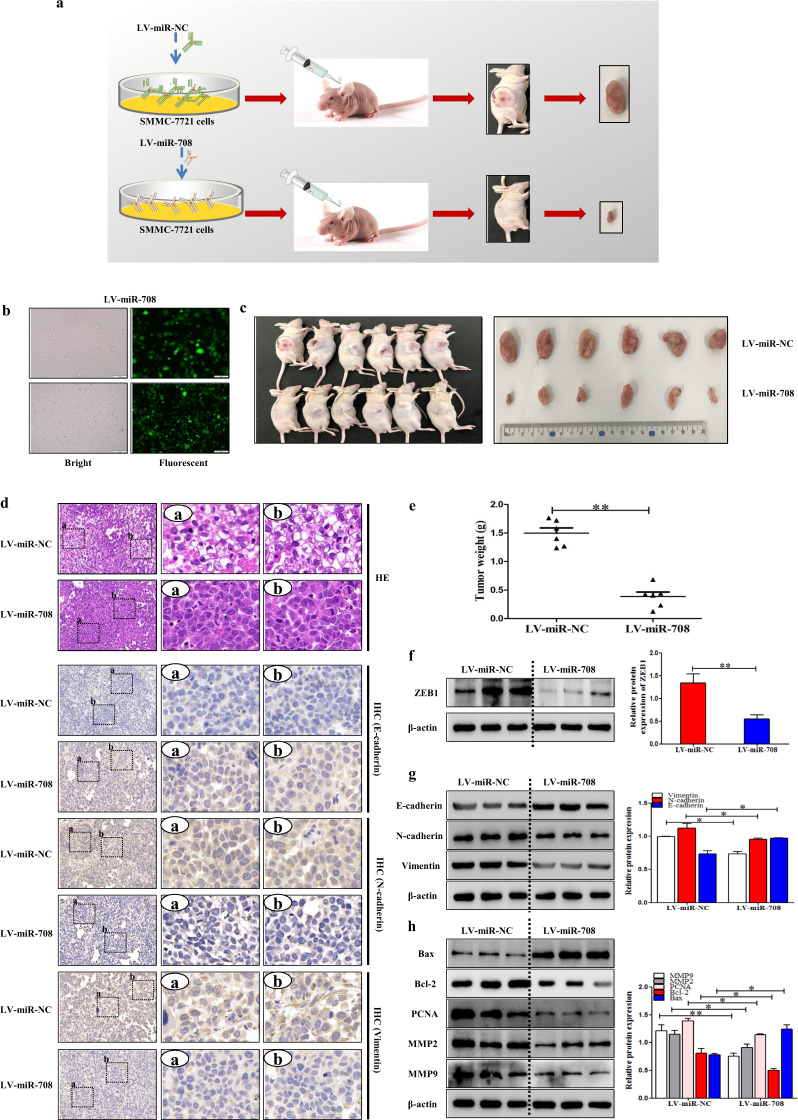
MiR-708 inhibited tumor growth in nude mice of HCC model
To detect the function of miR-708 on tumor formation in vivo, LV-miR-708 was used to establish a stable miR-708 overexpressing SMMC-7721 strain, and LV-miR-NC was used as a NC (Fig. 7a). First, LV-miR-708 was used for subcutaneous injection in nude mice, while LV-miR-NC was used as a control to establish a tumor model (Fig. 7b). As its diameter reached 1.0 cm, the tumors were excised from nude mice. the tumor was photographed and the volume was measured. The results showed that the tumor in the LV-miR-708 group grew much slower than the control group (Fig. 7c,e). The results of H&E staining showed that LV-miR-708 reduced the degree of HCC compared to LV-miR-NC (Fig. 7d). In addition, the IHC and Western blotting results showed that LV-miR-708 increased the expression level of E-cadherin and decreased the expression levels of Vimentin and N-cadherin (Fig. 7d,g), indicating that LV-miR-708 repressed the progress of EMT in nude mice of HCC model. Furthermore, the results of Western blotting demonstrated that LV-miR-708 inhibit HCC proliferation and apoptosis in nude mice (Fig. 7h), and the expression level of ZEB1 was decreased in nude mice injected with LV-miR-708-transfected cells compared to the mice injected with LV-miR-NC-transfected cells (Fig. 7f). Based on these observations, these results demonstrated that miR-708 could inhibit tumor growth in HCC model nude mice.
Fig. 7. MiR-708 inhibited tumor growth in nude mice of HCC model.
MiR-708 inhibited tumor growth in nude mice of HCC model. a A stable SMMC-7721 line was established by using LV-miR-708, with LV-miR-NC used as a negative control. Then, the HCC model was established by subcutaneous injection of nude mice of LV-miR-708 or control LV-miR-NC. b Stable control and LV-miR-708-transfected SMMC-7721 cell lines were observed by inversed fluorescent microscope. c, e Tumor images and weights at experimental endpoints in NC and LV-miR-708-transfected SMMC-7721 cells xenografts (n = 6 for each group). d The H&E stain in the LV-miR-NC group and LV-miR-708 group. IHC results of E-cadherin, N-cadherin, and Vimentin were shown in the LV-miR-NC group and LV-miR-708 group. f The protein expression of ZEB1 in the LV-miR-NC group and LV-miR-708 group. g Western Blotting analysis of E-cadherin, N-cadherin, and Vimentin in tumor tissues of LV-miR-NC, and LV-miR-708 groups. h Western blotting analysis of MMP9, MMP2, PCNA, Bax and Bcl-2 in tumor tissues of LV-miR-NC and LV-miR-708 groups. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
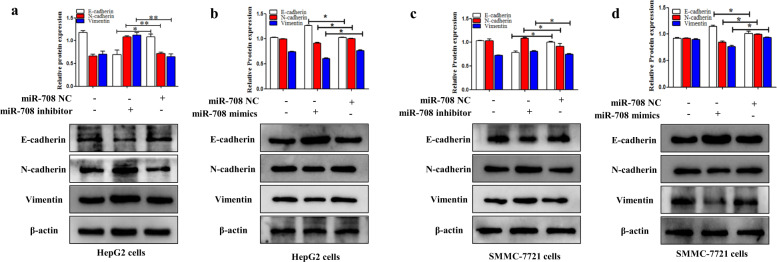
MiR-708 inhibited the process of EMT in HCC
The function of the miR-708 on the process of EMT was detected in HepG2 and SMMC-7721 cells. Furthermore, miR-708 mimics promoted the expression level of E-cadherin whereas inhibited the expression levels of N-cadherin and Vimentin (Fig. 8b,d). Notably, the function of miR-708 mimics on EMT could be reversed by miR-708 inhibitor (Fig. 8a,c). The data indicated that miR-708 could inhibit the process of EMT in HepG2 and SMMC-7721 cells.
Fig. 8. MiR-708 inhibited the process of EMT in HCC.
a–d The protein expression levels of E-cadherin, N-cadherin, and Vimentin were detected in HepG2 and SMMC-7721 cells transfected with miR-708 mimics and miR-708 inhibitor, respectively. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, compared to the control group.
MiR-708 inhibited HCC cells metastasis in vivo
To further validate the role of miR-708 in HCC metastasis, we performed animal experiments in nude mice. LV-miR-708 was used to establish a stable miR-708 overexpression SMMC-7721 cell line, and LV-miR-NC was used as a NC. First, a HCC metastatic model was established by stable miR-708 overexpression SMMC-7721 cell through tail-vein injection, while LV-miR-NC was used as a control group. Luciferase signals were detected using IVIS Lumina III Imaging on 30 min, 1 week, and 3 weeks after injection. Thirty minutes after injection, the tail-vein injection was confirmed by the distributing luminescence signal throughout the body of the mice (Fig. S4). Furthermore, results showed that HCC metastasis was inhibited in the LV-miR-708 group (Fig. S4). In mice of the LV-miR-NC group, the formation of metastatic lesions was first detected on 1 week and 3 weeks after tail-vein injection. The results demonstrated that HCC cell metastasis model showed that overexpression of miR-708 inhibited the metastasis of HCC cells compared to the control group. Therefore, miR-708 inhibited the metastasis of HCC cells.
Discussion
In recent years, the cognition of the mechanism of ZEB1 in cancer evolution has been largely developed [39]. ZEB1 was one such EMT-transcription factor, which was first reported in breast cancer [40, 41], and it was also involved in the progression of lung cancer, gastric cancer, and prostate cancer [42–44]. Our previous study showed that ZEB1 aggravated liver fibrosis [19]. More than 80% of HCC developed in fibrotic suggesting an important role of liver fibrosis in the premalignant environment of the liver [25]. HCC is one of the increasing major health problems in both developing and developed countries [45]. Here, this study will find the relationship between ZEB1 and HCC. In the present study, TCGA database analysis showed that the mRNA expression level of ZEB1 was upregulated in HCC tissues than in normal tissues. Furthermore, clinical data from TCGA database confirmed that HCC patients in the high-risk group had significantly worse prognosis than those in the low-risk group. We collected the HCC tissues from the First Affiliated Hospital of Anhui Medical University and the results of MRI proved that there were multiple low-density nodules and masses scattered in the liver parenchyma. H&E staining further demonstrated that there was a large area of necrosis in human HCC tissues. Notably, these results showed that the expression level of ZEB1 was significantly increased both in HCC tissues and HCC cell lines (HepG2 and SMMC-7721 cells).
MiRNAs could exert its biological function by mediating target genes [46]. MiR-708 is correlated with cell proliferation, migration, invasion, and EMT via targeting genes. For example, Wu et al. identified that miR-708 could increase apoptosis of synovial fibroblast MH7A cells and inhibit colony formation and migration [47]. Previous study found that miR-708 mimics decreased cell proliferation and invasion and promoted apoptosis in human glioblastoma [48]. Bioinformatics tools (TargetScan and miRanda) were used to predict the target genes of miR-708. The result found that ZEB1 may be a direct target of miR-708. Therefore, the 3′-UTR of ZEB1 was cloned into the pmirGLO luciferase reporter vector and co-transfected into HepG2 cells with miR-708 and miR-NC, respectively, to determine if miR-708 passed the corresponding 3′-UTR combines to adjust ZEB1. Dual luciferase reporter assay revealed that miR-708 significantly decreased ZEB1 luciferase activity in HepG2 cells. RT-qPCR and Western blotting analysis respectively confirmed ZEB1 as a functional target of miR-708. ZEB1 was identified as a target of miR-708 and is negatively regulated by miR-708 at the posttranscriptional level via a specific target site within the 3′-UTR of ZEB1. However, the expression and the potential biological function of miR-708 in HCC cells have not been fully elucidated. This study mainly found that miR-708 was significantly downregulated in HCC tissues and HCC cells (HepG2 and SMMC-7721 cells), while miR-708 mimics could increase HCC cell apoptosis. Moreover, ZEB1 inhibited HCC cell apoptosis by using Western blotting analysis and the flow cytometry assay. On the one hand, EdU staining and Western blotting were used to detect the function of miR-708 and ZEB1 on the cell proliferation of HCC, these results determined that miR-708 could inhibit HCC cell proliferation by targeting ZEB1. On the other hand, the results of Transwell migration and invasion assay suggested that miR-708 repressed HCC cell migration and invasion whereas ZEB1 promoted HCC cell migration and invasion, the results of Western blotting were consistent with it. Furthermore, miR-708 could inhibit the process of EMT both in human HCC tissues and cell lines by targeting ZEB1. In vivo, nude mice received subcutaneous injection of SMMC-7721 cells treated with LV-miR-708 or control LV-miR-NC to establish tumor models. The results demonstrated that the nude mice treated with LV-miR-708 significantly decreased the expression level of ZEB1 compared to that in the control group. Furthermore, we have performed animal experiments to validate that miR-708 can also inhibit metastasis in vivo. The results demonstrated that overexpression miR-708 inhibited HCC cell metastasis compared to the control group in HCC cell metastasis model. This approach provides novel insights and potential treatment strategies for the progress of HCC treatment. These data together indicated that miR-708 suppresses the metastasis and EMT of HCC cells through directly targeting ZEB1.
Mechanically, increasingly studies have demonstrated that the Wnt/β-catenin signaling pathway was involved in the development of a variety of human cancers, including colorectal cancer, breast cancer, prostate cancer, and HCC [49–53]. Various cellular functions, such as apoptosis, proliferation, migration, and invasion, were involved in Wnt-dependent carcinogenesis. The Wnt/β-catenin signaling pathway participated in the progression of HCC and malignant events such as the EMT, metastasis, and invasion [54]. These results showed that the Wnt/β-catenin signaling pathway has the latent prognosis and treatment value in HCC. The experiment implicated that miR-708 could inhibit HCC proliferation, migration, and invasion by inhibiting β-catenin through the Wnt/β-catenin signaling pathway, which could be proved by the downregulated protein level of β-catenin in HepG2 and SMMC-7721 cells. Furthermore, ZEB1 could increase the expression level of β-catenin. The target genes of the Wnt/β-catenin signaling pathway were c-Myc and cyclin D1 [55–57]. The results showed higher protein expression levels of cyclin D1 and c-Myc in HepG2 and SMMC-7721 cells in HCC tissues. Moreover, ZEB1 siRNA could weaken the expression levels of c-Myc and cyclin D1, indicating that ZEB1 might mediate HCC cell proliferation, migration, and invasion via the Wnt/β-catenin signaling pathway. Based on these observations, we could conclude that miR-708 inhibits HCC cells proliferation, migration/invasion via the Wnt/β-catenin signaling pathway by targeting ZEB1.
To summarize, the data indicated that ZEB1 is essential for the development of HCC regulated by miR-708 via the Wnt/β-catenin signaling pathway in HepG2 and SMMC-7721 cells. In future research, the study will focus on more features of ZEB1 in the development of HCC. Based on the function of ZEB1, research on targeted drugs of ZEB1 for HCC was subsequently carried out. Understanding the functions of ZEB1 on inflammation, autophagy, and other functions during HCC is a meaningful work, which can provide a bright future for the treatment of HCC.
Supplementary information
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Nos. 81700522, 81602344), the fund of Anhui Medical University doctoral start research (No. 0601067101), Anhui Provincial Natural Science Foundation (1704a0802161, 1808085MH235). The fund of Anhui Science and Technology Department Soft Science Project (1607a0202062). We would like to thank Dr Su-wen Li, Department of Gastroenterology, the First Affiliated Hospital of Anhui Medical University for providing us with patient medical records and liver samples.
Author contributions
LYL and JFY performed the cell experiment and analyzed the data. FR and ZPL performed the animal experiments. TX and JL designed, supervised and wrote the manuscript. SH, YW and HF provided a series of experimental instructions and help. WHK and RY do TCGA data analysis. XWF and BJC contributed new reagents or analytic tools. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Liang-yun Li, Jun-fa Yang, Fan Rong
Supplementary information
The online version of this article (10.1038/s41401-020-00575-3) contains supplementary material, which is available to authorized users.
References
- 1.Krishan S, Dhiman RK, Kalra N, Sharma R, Baijal SS, Arora A, et al. Joint Consensus Statement of the Indian National Association for Study of the Liver and Indian Radiological and Imaging Association for the Diagnosis and Imaging of Hepatocellular Carcinoma Incorporating Liver Imaging Reporting and Data System. J Clin Exp Hepatol. 2019;9:625–51.. doi: 10.1016/j.jceh.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaceanu LA. Biomarkers vs imaging in the early detection of hepatocellular carcinoma and prognosis. World J Clin Cases. 2019;7:1367–82.. doi: 10.12998/wjcc.v7.i12.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol. 2020;235:6929–41.. doi: 10.1002/jcp.29589. [DOI] [PubMed] [Google Scholar]
- 4.Yin L, Cai Z, Zhu B, Xu C. Identification of key pathways and genes in the dynamic progression of HCC based on WGCNA. Genes. 2018;9:92. doi: 10.3390/genes9020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reghupaty SC, Sarkar D. Current status of gene therapy in hepatocellular carcinoma. Cancers. 2019;11:1265. doi: 10.3390/cancers11091265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, Huang H, Yu L, Cao L. Meta-analysis of gene expression profiles indicates genes in spliceosome pathway are up-regulated in hepatocellular carcinoma (HCC) Med Oncol. 2015;32:96. doi: 10.1007/s12032-014-0425-6. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76.. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 8.Klungboonkrong V, Das D, McLennan G. Molecular mechanisms and targets of therapy for hepatocellular carcinoma. J Vasc Inter Radio. 2017;28:949–55.. doi: 10.1016/j.jvir.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73.. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42.. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 13.Zhai B, Yan HX, Liu SQ, Chen L, Wu MC, Wang HY. Reduced expression of E-cadherin/catenin complex in hepatocellular carcinomas. World J Gastroenterol. 2008;14:5665–73. doi: 10.3748/wjg.14.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fransvea E, Angelotti U, Antonaci S, Giannelli G. Blocking transforming growth factor-beta up-regulates E-cadherin and reduces migration and invasion of hepatocellular carcinoma cells. Hepatology. 2008;47:1557–66.. doi: 10.1002/hep.22201. [DOI] [PubMed] [Google Scholar]
- 15.Katsuyama E, Yan M, Watanabe KS, Narazaki M, Matsushima S, Yamamura Y, et al. Downregulation of miR-200a-3p, targeting CtBP2 complex, is involved in the hypoproduction of IL-2 in systemic lupus erythematosus-derived T cells. J Immunol. 2017;198:4268–76.. doi: 10.4049/jimmunol.1601705. [DOI] [PubMed] [Google Scholar]
- 16.Peng DH, Kundu ST, Fradette JJ, Diao L, Tong P, Byers LA, et al. ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci Transl Med. 2019;11:eaaq1238. doi: 10.1126/scitranslmed.aaq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Lee S, Teh CE, Bunting K, Ma L, Shannon MF. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int Immunol. 2009;21:227–35.. doi: 10.1093/intimm/dxn143. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Liu YM, Chen C, Li LY, Zhang BY, Yang JF, et al. MicroRNA-708 prevents ethanol-induced hepatic lipid accumulation and inflammatory reaction via direct targeting ZEB1. Life Sci. 2020;258:118147. doi: 10.1016/j.lfs.2020.118147. [DOI] [PubMed] [Google Scholar]
- 19.Li LY, Yang CC, Yang JF, Li HD, Zhang BY, Zhou H, et al. ZEB1 regulates the activation of hepatic stellate cells through Wnt/beta-catenin signaling pathway. Eur J Pharmacol. 2019;865:172787. doi: 10.1016/j.ejphar.2019.172787. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, Lee CH, Lee SW. Hepatitis C virus infection stimulates transforming growth factor-beta1 expression through up-regulating miR-192. J Microbiol. 2016;54:520–6. doi: 10.1007/s12275-016-6240-3. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee R, Mitra A. An overview of effective therapies and recent advances in biomarkers for chronic liver diseases and associated liver cancer. Int Immunopharmacol. 2015;24:335–45.. doi: 10.1016/j.intimp.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 22.Adachi Y, Takeuchi T, Nagayama T, Ohtsuki Y, Furihata M. Zeb1-mediated T-cadherin repression increases the invasive potential of gallbladder cancer. FEBS Lett. 2009;583:430–6. doi: 10.1016/j.febslet.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 23.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–29.. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 24.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30.. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Affo S, Yu LX, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–86.. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baglieri J, Brenner DA, Kisseleva T. The role of fibrosis and liver-associated fibroblasts in the pathogenesis of hepatocellular carcinoma. Int J Mol Sci. 2019;20:1723. doi: 10.3390/ijms20071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Q, Yu DY, Yang ZF, Liu DY, Cao HQ, Liao XW. IGFBP2 upregulates ZEB1 expression and promotes hepatocellular carcinoma progression through NF-kappaB signaling pathway. Dig Liver Dis. 2020;52:573–81.. doi: 10.1016/j.dld.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez-Mateos EM. Role of MicroRNAs in innate neuroprotection mechanisms due to preconditioning of the brain. Front Neurosci. 2015;9:118. doi: 10.3389/fnins.2015.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caviglia JM, Yan J, Jang MK, Gwak GY, Affo S, Yu L, et al. MicroRNA-21 and dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology. 2018;67:2414–29.. doi: 10.1002/hep.29627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong XY, Zhang Y. Protective effect of miR-20a against hypoxia/reoxygenation treatment on cardiomyocytes cell viability and cell apoptosis by targeting TLR4 and inhibiting p38 MAPK/JNK signaling. Vitr Cell Dev Biol Anim. 2019;55:793–800. doi: 10.1007/s11626-019-00399-4. [DOI] [PubMed] [Google Scholar]
- 31.Liao W, Zhang Y. MicroRNA-381 facilitates autophagy and apoptosis in prostate cancer cells via inhibiting the RELN-mediated PI3K/AKT/mTOR signaling pathway. Life Sci. 2020;254:117672. doi: 10.1016/j.lfs.2020.117672. [DOI] [PubMed] [Google Scholar]
- 32.Ni X, Lin Z, Dai S, Chen H, Chen J, Zheng C, et al. Screening and verification of microRNA promoter methylation sites in hepatocellular carcinoma. J Cell Biochem. 2020;121:3626–41.. doi: 10.1002/jcb.29656. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Tao Q, Zhou Y, Chen Q, Li L, Hu S, et al. MicroRNA-708 represses hepatic stellate cells activation and proliferation by targeting ZEB1 through Wnt/beta-catenin pathway. Eur J Pharmacol. 2020;871:172927. doi: 10.1016/j.ejphar.2020.172927. [DOI] [PubMed] [Google Scholar]
- 34.Pan LX, Li LY, Zhou H, Cheng SQ, Liu YM, Lian PP, et al. TMEM100 mediates inflammatory cytokines secretion in hepatic stellate cells and its mechanism research. Toxicol Lett. 2019;317:82–91. doi: 10.1016/j.toxlet.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Xu T, Pan LX, Ge YX, Li P, Meng XM, Huang C, et al. TMEM88 mediates inflammatory cytokines secretion by regulating JNK/P38 and canonical Wnt/beta-catenin signaling pathway in LX-2 cells. Inflammopharmacology. 2018;26:1339–48.. doi: 10.1007/s10787-017-0419-z. [DOI] [PubMed] [Google Scholar]
- 36.Hou FJ, Guo LX, Zheng KY, Song JN, Wang Q, Zheng YG. Chelidonine enhances the antitumor effect of lenvatinib on hepatocellular carcinoma cells. Onco Targets Ther. 2019;12:6685–97.. doi: 10.2147/OTT.S215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin XL, Liu M, Liu Y, Hu H, Pan Y, Zou W, et al. Transforming growth factor beta1 promotes migration and invasion in HepG2 cells: Epithelialtomesenchymal transition via JAK/STAT3 signaling. Int J Mol Med. 2018;41:129–36. doi: 10.3892/ijmm.2017.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B, Zhou S, Zhan Y, Ke J, Wang K, Liang Q, et al. Dioscin inhibits the invasion and migration of hepatocellular carcinoma HepG2 cells by reversing TGF-beta1-induced epithelial-mesenchymal transition. Molecules. 2019;24:2222. doi: 10.3390/molecules24122222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HT, Zhong HT, Li GW, Shen JX, Ye QQ, Zhang ML, et al. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J Transl Med. 2020;18:51. doi: 10.1186/s12967-020-02240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Zhou C, Jiang H, Liang L, Shi W, Zhang Q, et al. ZEB1 induces ER-alpha promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8:e2732. doi: 10.1038/cddis.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar A, Rahaman A, Biswas I, Mukherjee G, Chatterjee S, Bhattacharjee S, et al. TGFbeta mediated LINC00273 upregulation sponges mir200a-3p and promotes invasion and metastasis by activating ZEB1. J Cell Physiol. 2020;235:7159–72.. doi: 10.1002/jcp.29614. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z, et al. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26:527–34.. doi: 10.1016/j.suronc.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–35.. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su W, Xu M, Chen X, Chen N, Gong J, Nie L, et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim DW, Talati C, Kim R. Hepatocellular carcinoma (HCC): beyond sorafenib-chemotherapy. J Gastrointest Oncol. 2017;8:256–65.. doi: 10.21037/jgo.2016.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng H, Ma R, Wang Q, Zhang P, Li D, Wang Q, et al. MiR-625-3p promotes cell migration and invasion via inhibition of SCAI in colorectal carcinoma cells. Oncotarget. 2015;6:27805–15.. doi: 10.18632/oncotarget.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Fan W, Ma L, Geng X. miR-708-5p promotes fibroblast-like synoviocytes’ cell apoptosis and ameliorates rheumatoid arthritis by the inhibition of Wnt3a/beta-catenin pathway. Drug Des Devel Ther. 2018;12:3439–47.. doi: 10.2147/DDDT.S177128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo P, Lan J, Ge J, Nie Q, Mao Q, Qiu Y. miR-708 acts as a tumor suppressor in human glioblastoma cells. Oncol Rep. 2013;30:870–6. doi: 10.3892/or.2013.2526. [DOI] [PubMed] [Google Scholar]
- 49.Xu Q, Krause M, Samoylenko A, Vainio S. Wnt signaling in renal cell carcinoma. Cancers. 2016;8:57. doi: 10.3390/cancers8060057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/beta-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–81.. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 51.Schneider JA, Logan SK. Revisiting the role of Wnt/beta-catenin signaling in prostate cancer. Mol Cell Endocrinol. 2018;462:3–8. doi: 10.1016/j.mce.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Li B, Zhou L, Yu S, Su Z, Song J, et al. Prodigiosin inhibits Wnt/beta-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci U S A. 2016;113:13150–5. doi: 10.1073/pnas.1616336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilchez V, Turcios L, Marti F, Gedaly R. Targeting Wnt/beta-catenin pathway in hepatocellular carcinoma treatment. World J Gastroenterol. 2016;22:823–32. doi: 10.3748/wjg.v22.i2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang J, Qu Q, Guo Y, Xiang Y, Feng D. Tankyrases/beta-catenin signaling pathway as an Anti-proliferation and anti-metastatic target in hepatocarcinoma cell lines. J Cancer. 2020;11:432–40.. doi: 10.7150/jca.30976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janssens N, Andries L, Janicot M, Perera T, Bakker A. Alteration of frizzled expression in renal cell carcinoma. Tumour Biol. 2004;25:161–71.. doi: 10.1159/000081098. [DOI] [PubMed] [Google Scholar]
- 56.Furge KA, Chen J, Koeman J, Swiatek P, Dykema K, Lucin K, et al. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res. 2007;67:3171–6. doi: 10.1158/0008-5472.CAN-06-4571. [DOI] [PubMed] [Google Scholar]
- 57.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.