Abstract
Background & Aims
In colorectal cancer, approximately 95% of patients are refractory to immunotherapy because of low antitumor immune responses. Therefore, there is an exigent need to develop treatments that increase antitumor immune responses and decrease tumor burden to enhance immunotherapy.
Methods
The gut microbiome has been described as a master modulator of immune responses. We administered the human commensal, Lactobacillus rhamnosus GG (LGG), to mice and characterized the changes in the gut immune landscape. Because the presence of lactobacilli in the gut microbiome has been linked with decreased tumor burden and antitumor immune responses, we also supplemented a genetic and a chemical model of murine intestinal cancer with LGG. For clinical relevance, we therapeutically administered LGG after tumors had formed. We also tested for the requirement of CD8 T cells in LGG-mediated modulation of gut tumor burden.
Results
We detected increased colonic CD8 T-cell responses specifically in LGG-supplemented mice. The CD8 T-cell induction was dependent on dendritic cell activation mediated via Toll-like receptor-2, thereby describing a novel mechanism in which a member of the human microbiome induces an intestinal CD8 T-cell response. We also show that LGG decreased tumor burden in the murine gut cancer models by a CD8 T-cell–dependent manner.
Conclusions
These data support the potential use of LGG to augment antitumor immune responses in colorectal cancer patients and ultimately for increasing the breadth and efficacy of immunotherapy.
Keywords: Probiotics, Microbiome, LGG, Cancer, CD8 T Cells, Dendritic Cells, CRC
Abbreviations used in this paper: BC, Bacillus cereus; BMDC, bone-marrow–derived dendritic cell; CFSE, carboxyfluorescein succinimidyl ester; CRC, colorectal cancer; CXCL, chemokine ligands; DC, dendritic cell; DSS-AOM, dextran sulfate sodium–azoxymethane; FBS, fetal bovine serum; FCM, flow cytometry; HBSS, Hank’s buffered salt solution; IL, interleukin; LGG, Lactobacillus rhamnosus GG; mLn, mesenteric lymph node; MSH, MutS Homolog; MSI, microsatellite instability; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PD-1, anti programmed cell death protein 1; TLR, Toll-like receptor; WT, wild-type
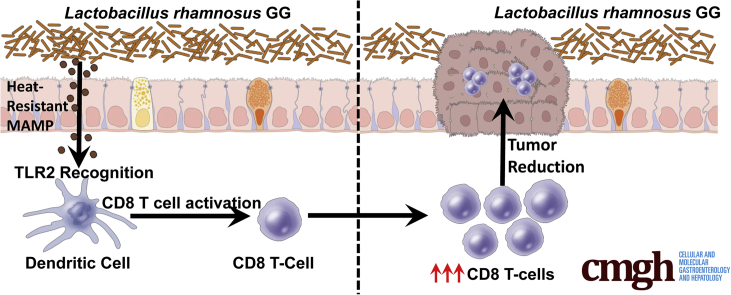
Graphical abstract
Summary.
Lactobacillus rhamnosus GG activates colonic CD8 T cells through a novel Toll-like receptor 2:dendritic cell axis. This induction of CD8 T cells could be used as a therapeutic modality to decrease tumor burden and treat colorectal cancer.
More than 1700 Americans per day died of a cancer-related death in 2019.1 Of these deaths, colorectal cancer (CRC) is the second highest cause of cancer-related mortality in the United States, although the incidence and mortality rates have stabilized over the past few years.2 However, globally, the incidence and mortality of CRC is increasing and cases of CRC are predicted to increase by 60% by 2030.3 With this negative outlook, innovative approaches and improvements to currently approved therapeutics are needed for enhanced treatment of CRC.
Recently, the advent of immunotherapy has revolutionized the fight against cancer. However, for CRC, only 10% of patients are approved for immunotherapy and more than 50% of these patients are refractory to treatment.4,5 The major determinant for CRC patients being approved for immunotherapy is microsatellite instability (MSI). This approval is because MSI-high tumors have increased levels of genomic instability. Genomic instability leads to abundant mutations, which culminates in a high output of tumor neoantigens. Because of increased tumor neoantigens, MSI-high tumors have greater immune cell infiltrate compared with their MSI-low counterparts. Tumors that have a heightened immune cell infiltrate correlate with greater responses in immunotherapy and better clinical outcomes.6, 7, 8, 9, 10, 11, 12 Therefore, one strategy for CRC treatment involves triggering MSI-low tumors to develop higher antitumor immune cell infiltrate such as their MSI-high counterparts. On the other hand, the immune cell infiltrate within MSI-high tumors can reach a point at which they no longer effectively clear tumor cells, reaching a state known as exhaustion.13 These exhausted immune cells are targeted in immunotherapy to restore their cytotoxic function and trigger antitumor immune responses. Unfortunately, immunotherapy response rates in MSI-high tumors still remain approximately 50%.4,5 Therefore, a critical challenge exists to increase the percentage of patients who respond to immunotherapy intervention.
Recently, the gut microbiome community structure was linked to immunotherapy response rates.14, 15, 16 Interestingly, the microbiome of nonresponding immunotherapy patients lack lactobacilli, whereas lactobacilli were shown to be present in the microbiome of responding patients.15 In addition, other studies that investigated the influence of Lactobacillus rhamnosus GG (LGG) on gut health and disease have reported that LGG and other lactobacilli promote an anti-inflammatory response in the intestine by regulating interleukin (IL)10 levels and promoting regulatory T-cell activity.17, 18, 19 At first, these reports seemed contradictory because increasing immunotherapy response rates would imply proinflammatory responses and modulating regulatory T cells would suggest anti-inflammatory responses. However, LGG also was reported to modulate the immune landscape in a proinflammatory manner, including increased M1 macrophage polarization, increased antibody production, modulation of dendritic cell activity, and reducing viral burden during influenza infections.20, 21, 22, 23 Because of these data, we hypothesized that members of the gut microbiome, specifically lactobacilli such as Lactobacillus rhamnosus GG, may hold promise in augmenting colonic immune responses and colonic antitumor responses.
To date, clinically plausible approaches to modulate the microbiome to induce increased CD8 T-cell infiltration into tumors, and by extension decreased tumor burden, remains to be explored thoroughly. In addition, the cell and molecular signaling cascades that the microbiome triggers within gut mucosal immune cells still is largely unclear. To address this gap in knowledge, we investigated the extent to which the human commensal, LGG, could modulate CD8 T-cell responses, the mechanism by which this occurs, and the therapeutic efficacy of this commensal bacterium in reducing colonic tumors in an immune-mediated manner.
Herein, we show that supplementation of mice with LGG expands the prevalence of CD8 T cells in the colonic mucosa. This expansion results through signaling cascades dependent on Toll-like receptor-2 (TLR2) on dendritic cells. We show that LGG administered as a therapeutic to colonic tumors can drive CD8 T cells to initiate antitumor immune responses. This response was dependent on CD8 T cells for greatest attenuation of colonic tumor burden. Therefore, we show that LGG can induce CD8 T-cell–driven antitumor immune responses and show the potential of LGG holding clinical promise as a co-therapeutic to be administered in conjunction with currently approved immunotherapy.
Results
LGG Orchestrates a Colonic CD8 T-Cell Response
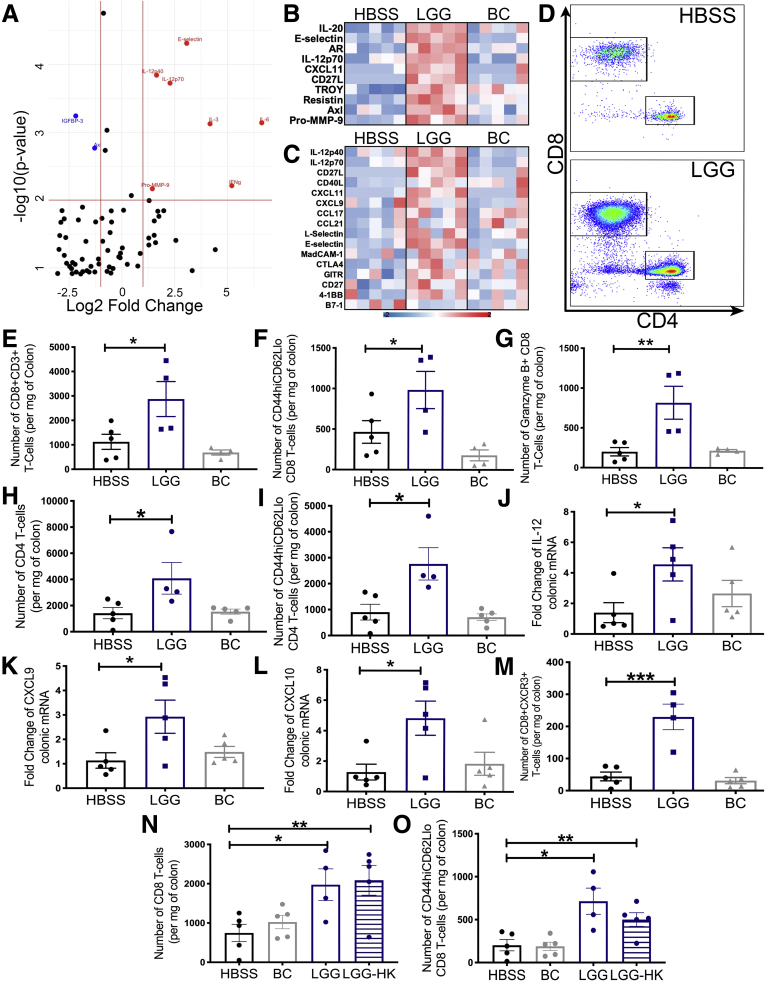
Because LGG has been reported to have both proinflammatory and anti-inflammatory properties, we first investigated if oral administration of LGG could modulate the murine colonic immune response by performing an expansive antibody array. We administered either LGG, a control nonprobiotic gram-positive bacterium known as Bacillus cereus (BC) or a vehicle control of Hank’s buffered salt solution (HBSS) daily by oral gavage for 2 weeks to 6-week-old specific pathogen-free C57BL/6 mice, and then performed an antibody array on protein extracted from total colonic tissue. Specifically, in LGG-treated mice, we detected several increased levels of several cytokines including IL12 and interferon-γ (Figure 1A). We also detected altered levels of chemokine ligands (CXCL) such as CXCL11 specifically in LGG-treated mice, which pointed to an altered immune response and potentially a modified T-cell response (Figure 1B and C). Because several reports exist of anti-inflammatory properties of LGG, we wanted to confirm these findings and elucidate LGG’s effect on the immune response in greater detail. Therefore, we took another cohort of C57BL/6 mice and administered either LGG, BC, or HBSS daily for 1 week. Mice were killed and the colonic lamina propria lymphocytes were analyzed by flow cytometry (FCM). We detected a significant increase in total numbers of CD8 T cells as well as effector and cytotoxic CD8 T cells in LGG-treated mice (Figure 1D–G). Furthermore, we detected an increase in total CD4 T cells and CD4 effector T cells, indicating that both subsets increased with LGG treatment (Figure 1H and I). In addition, analysis for transcript enrichment of chemokines and cytokines by LGG as observed in the antibody array confirmed a significant increase in IL12 and chemokines that function in CD8 T-cell trafficking, namely CXCL9/10. (Figure 1J–L). Similar to the increase in CXCL9/10, we detected an increase in the number of colonic CXCR3+ CD8 T cells that bind CXCL9/10 in LGG-treated mice (Figure 1M). Importantly, we did not observe any significant changes in CD8 T-cell numbers in the spleens of LGG-treated mice (data not shown), showing that LGG only induces a CD8 T-cell immune response localized to the gut. To determine if a specific element from live LGG is necessary to elicit the CD8 T-cell response, we administered LGG that was heat-killed for 10 minutes at 60°C to mice for 1 week. We found that heat-killed LGG also was able to increase total colonic CD8 T cells and effector CD8 T cells (Figure 1N and O). These data establish that LGG is capable of inducing colonic CD8 T cells and this induction occurs even in response to heat-killed LGG.
Figure 1.
Lactobacillus rhamnosus GG orchestrates a colonic CD8 T-cell response. (A) Specific pathogen-free C57BL/6J mice were supplemented with HBSS, BC, or LGG, or by oral gavage daily for 2 weeks before colons were harvested and analyzed for protein abundance using the Quantibody Mouse Cytokine Array 4000 (RayBiotech, Inc). A volcano plot representation is shown of cytokines that were significantly more abundant in colonic tissues of LGG-supplemented mice compared with colons of mice supplemented with BC. Axes represent log2 fold change (x-axis) against -log10 (P value) (y-axis). (B) Heatmap representation of the top 10 differentially abundant proteins in LGG mice compared with HBSS-supplemented mice detected in the colonic tissue of mice described in panel A. (C) Heatmap representation of the top 16 differentially expressed immune-mediated proteins detected in the colonic tissue of the 3 groups. (D) Flow cytometry analysis for the detection of immune cells in the colonic epithelium of C57BL/6J mice administered either HBSS, LGG, or BC for 1 week. Representative FCM plots shown of T cells. (E) Quantification of CD8 T cells in panel D. (F) Quantification of effector CD8 T cells in panel D. (G) Quantification of granzyme-B–expressing CD8 T cells in panel D. (H) Quantification of CD4 T cells in panel D. (I) Quantification of effector CD4 T cells. (J) Reverse-transcription PCR analysis for the detection of il-12 transcripts in the colonic tissue of mice in panel D. (K) Detection of cxcl9 transcripts in the colonic tissue of mice in panel D. (L) Detection of cxcl10 transcripts in the colonic tissue of mice in panel D. (M) Quantification of CXCR3+ CD8 T cells for analysis described in panel D. (N) Flow cytometry analysis for the detection of immune cells in the colonic epithelium of C57BL/6J mice administered either HBSS, BC, LGG, or heat-killed (HK) LGG for 1 week. The chart represents quantification of the number of CD8 T cells in the colonic tissues of these mice. (O) Quantification of effector CD8 T cells in mice described in panel L. Statistical significance was tested by 1-way analysis of variance for all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. n = 4/5 for all experiments. mRNA, messenger RNA.
LGG Primes and Requires Dendritic Cells for Increased CD8 T-Cell Responses
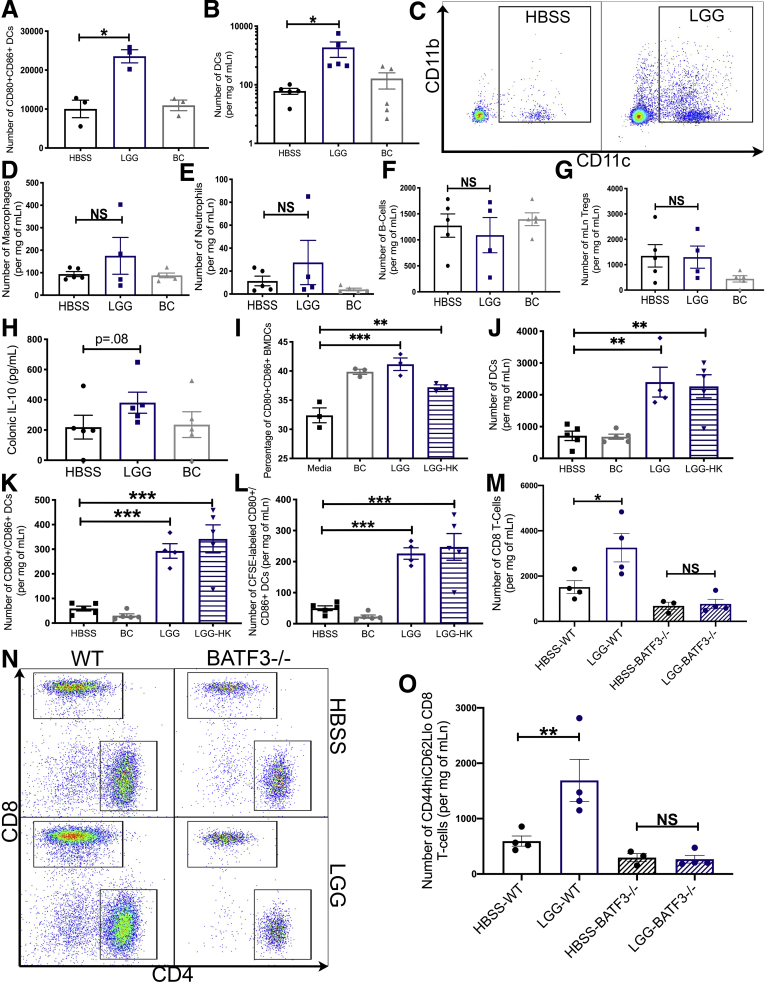
A previous study showed that LGG could activate dendritic cells (DCs) in vitro.21 We first corroborated this finding by showing that LGG can activate bone-marrow–derived DCs (BMDCs) in vitro (Figure 2A). Because DCs function in the expansion of CD8 T cells, we investigated whether this response to LGG also occurs in vivo. We administered either LGG, BC, or HBSS daily for 1 week to C57BL/6 mice and isolated the mesenteric lymph node (mLn), which collects the lymphatic fluid from the colon and the small intestine. We detected a significant increase in the total number of DCs in the mLn of LGG-treated mice (Figure 2B and C). However, we did not detect changes to the number of other immune cells including macrophages, neutrophils, and B cells in LGG-treated mice (Figure 2D–F). In addition, we did not detect changes to the numbers of regulatory T cells and did not detect a significant increase in IL10 levels (Figure 2G and H). Although we did not further characterize these cell types, it is noteworthy that other reports have shown that LGG can influence the activation of these cells.23, 24, 25 Because heat-killed LGG could activate CD8 T cells, we tested whether heat-killed LGG was capable of inducing changes to DCs in vitro and in vivo. First, we generated BMDCs from C57BL/6 mice and incubated them with either HBSS, BC, LGG, or heat-killed LGG. FCM analysis detected significant DC activation after incubation with both LGG and heat-killed LGG (Figure 2I). Importantly, we investigated how LGG was activating DCs in vivo and whether direct contact with LGG by surveilling gut DCs was required. To determine this, in a new cohort of C57BL/6 mice, we administered either HBSS or fluorescently tagged (carboxyfluorescein succinimidyl ester [CFSE]) BC, LGG, or heat-killed LGG. CFSE covalently links with lysine residues and other amines, effectively labeling the bacteria. This approach would allow tagging and tracking of DCs that had sampled and taken up the CFSE-labeled bacteria after the DCs had egressed from the gut to the mLn. Analysis of treated mLns by FCM detected both LGG and heat-killed LGG, increased DC numbers, and activation (Figure 2J and K). Critically, LGG and heat-killed LGG activated DCs were positive for CFSE, indicating that these DCs indeed had interacted with LGG before trafficking to the mLn (Figure 2L). Based on these data, we hypothesized that LGG requires DCs to increase CD8 T-cell numbers in the colon. Therefore, to test this, we treated a cohort of wild-type (WT) C57BL/6 or BATF3-/- mice that lack conventional DCs with HBSS or LGG and found that LGG did not increase the number of CD8 T cells in BATF3-/- mice compared with WT controls (Figure 2M–O). Altogether, these data support the conclusion that LGG drives CD8 T-cell expansion and responses via a DC-dependent mechanism.
Figure 2.
Lactobacillus rhamnosus GG primes and requires DCs for enhanced CD8 T-cell responses. (A) Detection by flow cytometry of the number of CD80+/CD86+ DCs in C57BL/6 mouse BMDCs after incubation with either HBSS, BC, or LGG for 24 hours. (B) Quantification of CD11B+ and CD11C+ DCs in the mLn of C57BL/6J mice supplemented for 1 week with either HBSS, LGG, or BC. (C) Representative flow cytometry analysis chart for experiment described in panel B. (D) Quantification of macrophages in mLn from mice in panel B. (E) Quantification of neutrophils in mLn from mice in panel B. (F) Quantification of B cells in mLn from mice in panel B. (G) Quantification of regulatory T cells in mLn from mice in panel B. (H) Colonic IL10 expression as determined by antibody array from Figure 1A–C. (I) Frequency of CD80+/CD86+ DCs in C57BL/6 mouse BMDCs after incubation with either HBSS, BC, LGG, or heat killed (HK) LGG for 24 hours. (J) Quantification of CD11b and CD11c+ DCs in the mLn of C57BL/6J mice supplemented for 1 week with CFSE-labeled HBSS, BC, LGG, or LGG-HK. (K) Number of CD80+CD86+ DCs in mLn of mice described in panel E. (L) Number of CFSE+CD80+CD86+ DCs in mLn of mice described in panel E. (M) Flow cytometry analysis for the detection of CD8 T cells in the mLn of B6.129S(C)-Batf3tm1Kmm/J (BATF3-/-) or C57BL/6 (WT) mice administered either HBSS or LGG by oral gavage for 1 week. (N) Representative flow cytometry analysis of mice described in panel H. (O) Quantification of effector CD8 T cells of mice described in panel H. Statistical significance was tested by 1-way analysis of variance (ANOVA) for all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. n = 3 for all BMDC experiments. (B) n = 5, (D–L) n = 4/5, and (M–O) n = 3/4. mRNA, messenger RNA.
LGG Requires TLR2 Signaling for DC Priming and CD8 T-Cell Activation
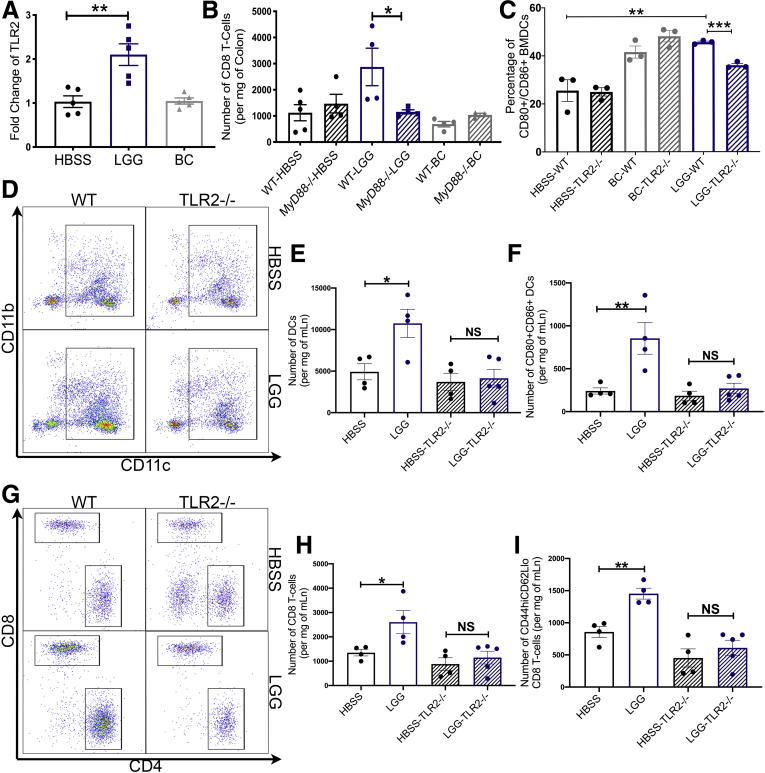
Because we had determined that DCs were required for LGG’s effect on CD8 T cells and the observation that heat-killed LGG still retained the effect, we investigated which elements within DCs are involved in sensing LGG and activating DCs. Previous reports have shown that some of LGG’s effect on the colonic epithelium occurred via TLR2 signaling.20,22, 26 Corroborating these reports, we detected transcript enrichment of tlr2 in colonic tissue of mice fed LGG (Figure 3A). To first test the requirement of TLRs for LGG-induced CD8 T-cell expansion, we administered LGG, BC, or HBSS daily to MyD88-/- mice for 1 week. Analysis of colonic lamina propria lymphocytes detected no changes in total CD8 T-cell numbers in MyD88-/- mice in response to LGG compared with WT controls (Figure 3B). This suggests that MyD88 indeed is required for LGG-induced CD8 T-cell expansion. However, many TLRs and even IL1 family cytokines can signal through MyD88. Thus, to test if TLRs, and specifically TLR2, were required, we generated BMDCs from both WT and TLR2-/- mice and incubated them with LGG, BC, or HBSS for 24 hours. We detected increased activation of WT BMDCs with LGG incubation, but this effect was abrogated in TLR2-/- BMDCs (Figure 3C). To elucidate this further, we administered LGG or HBSS daily to TLR2-/- or WT C57BL/6 mice for 1 week. Analysis by FCM showed no changes to the number or activation of DCs or changes to total or effector CD8 T-cell numbers in LGG-treated TLR2-/- mice (Figure 3D–I). Altogether, these data point to the conclusion that LGG is sensed by TLR2 on DCs, which thereafter increases DC number and activation, culminating in the activation of a CD8 T-cell response.
Figure 3.
Lactobacillus rhamnosus GG requires TLR2 signaling for DC priming and activation of CD8 T cells. (A) Reverse-transcription PCR analysis for the detection of tlr-2 transcripts in the colonic tissue of C57BL/6 mice supplemented with HBSS, BC, or LGG, or by oral gavage daily for 2 weeks. (B) Flow cytometry for the detection of CD8 T cells in the colons from C57Bl/6 (WT) or B6.129P2(SJL)-Myd88tm1.1Defr/J (Myd88-/-) mice administered either HBSS, LGG, or BC by oral gavage for 1 week. (C) Detection of CD80+/CD86+ BMDCs derived from either WT (C57BL/6) mice, or from TLR2-/- (B6.129-Tlr2tm1Kir/J) mice after incubation with either HBSS, BC, or LGG for 24 hours. (D) Representative flow cytometry plots from the analysis of mLns from WT (C57BL/6) mice, or from TLR2-/- (B6.129-Tlr2tm1Kir/J) mice supplemented with HBSS or LGG for 1 week. (E) Quantification of DCs in the mLn of mice described in panel C. (F) Number of CD80+CD86+ DCs in mLns of mice described in panel C. (G) Quantification of T cells in mice described in panel C. (H) Quantification of CD8 T cells in mice described in panel C. (I) Quantification of effector CD8 T cells in mice described in panel C. One-way analysis of variance was used for statistical analysis and represented as ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (A) n = 5, (C) n = 3; for MyD88-/- and TLR2-/- experiments, n = 4–5.
LGG Supplementation Decreases Colonic Tumor Burden in a Genetic Cancer Model
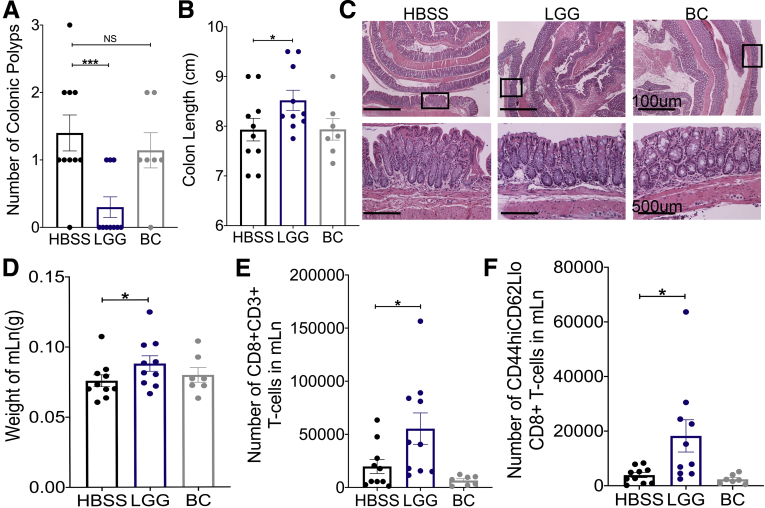
In CRC, the CD8 T-cell immune response performs a critical function both in MSI-high colonic tumors, and in inherited Lynch syndrome CRC.27 To examine if LGG supplementation influences colonic tumor burden in these contexts, we used a model of Lynch syndrome in which intestinal-specific MutS Homolog 2 (MSH2) knockout mice are generated.28 Both male and female intestinal-specific MSH2 knockout mice were raised until 7 months of age and then subjected to 6 weeks of either HBSS, LGG, or BC supplementation. After 6 weeks, LGG-supplemented animals showed significantly lower colonic tumor burden compared with HBSS- or BC-supplemented mice (Figure 4A). In addition, colons of LGG-treated mice were significantly longer than controls (Figure 4B). However, total tumor counts generally were low in the MSH2 model, and overall histologic analysis by H&E could not detect significant changes in LGG-treated mice, probably owing to the low tumor burden present (Figure 4C). Nevertheless, we detected significantly increased mLn weight and increased total and effector CD8 T cells in the mLn of LGG-supplemented mice compared with controls (Figure 4D–F). Together, these data illustrated that LGG expanded the CD8 T-cell population and significantly reduced tumor burden in a genetic model of intestinal cancer.
Figure 4.
LGG supplementation decreases colonic tumor burden in a genetic cancer model. (A) Intestinal-specific MSH2 knockout mice were generated by crossing Villin-Cre mice with MSH2loxP until homozygosity. Intestinal-specific MSH2 knockout mice were raised to 7 months of age, whereupon they were supplemented with HBSS, LGG, or BC by oral gavage for 6 weeks. After 6 weeks of supplementation, quantification of tumor burden was determined after removal and opening the colon longitudinally. (B) Colon lengths of mice described in panel A measured from rectum to cecum before removing colonic contents or any other manipulation. (C) H&E sections of Swiss-rolled colonic tissue with representative images of low-powered (upper panels) and high-powered (lower panels) views. Areas within lower panels indicated by rectangle within the upper panels. (D) Weights of mLn in mice described in panel A. (E) Flow cytometry analysis for the detection total CD8 T-cell numbers in mLn of mice described in panel A. (F) Quantification of total effector CD8 T cells from samples described in panel E. One-way analysis of variance was used for statistical significance with ∗P < .05 and ∗∗∗P < .001; n = 10, 10, and 7 for HBSS, LGG, and BC, respectively. Groups were made up of approximately half male and female mice with 5, 5, and 3 male mice for HBSS, LGG, and BC, respectively.
Therapeutic LGG Administration Attenuates Tumors in a Chemical Colonic Cancer Model
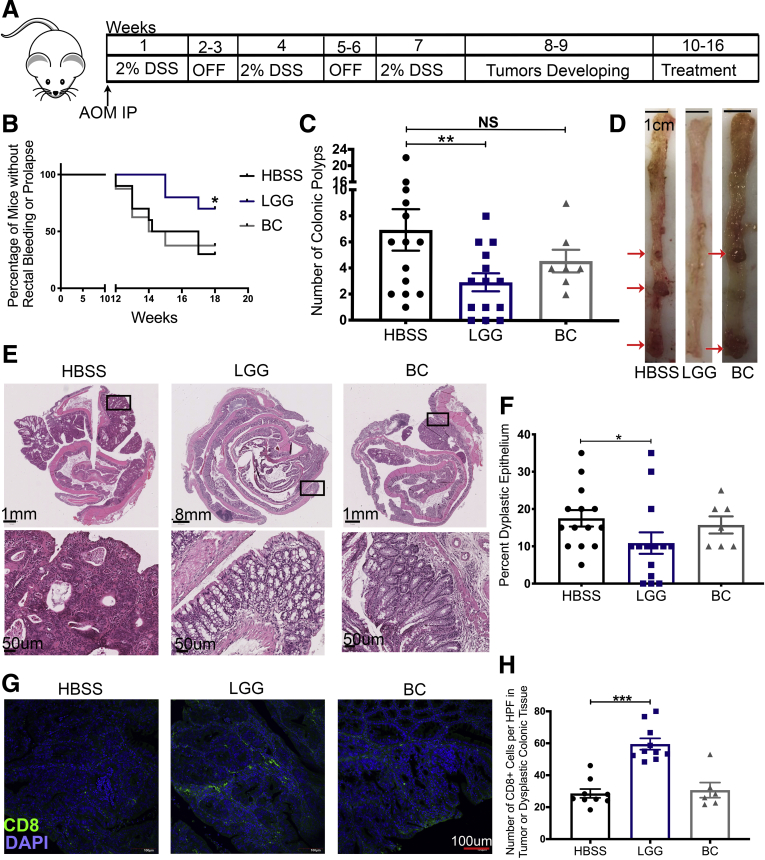
Because the intestinal-specific MSH2 knockout mice did not generate many tumors, we deemed that the model was not optimal to examine LGG-induced modulation of T-cell infiltration into tumors. As an alternative and complementary approach, we induced colonic tumors in mice through the well-established dextran sulfate sodium–azoxymethane (DSS-AOM) chemical model of colitis-associated carcinoma. Previous studies on LGG in CRC models have always administered the bacteria prophylactically.29, 30, 31, 32 By contrast, we administered LGG, BC, or HBSS daily after tumors had been established as outlined in the experimental design (Figure 5A). In LGG-treated animals, we observed significantly lower clinical manifestation of disease including less rectal bleeding or rectal prolapse (Figure 5B). At death, we observed fewer visible colonic tumors in LGG-treated mice (Figure 5C and D). Furthermore, H&E staining and subsequent histologic analysis showed less dysplastic epithelium in LGG-treated colons (Figure 5E and F). Importantly, immunofluorescent analysis for CD8 T cells detected significantly more intratumoral or intradysplastic epithelial CD8 T cells in LGG-treated samples as compared with controls showing an LGG-induced boost to cytotoxic CD8 T-cell antitumor response (Figure 5G and H). Together, these data show that therapeutic administration of LGG reduced tumor burden in a mouse model of CRC and induced the recruitment of CD8 T cells into the tumor microenvironment.
Figure 5.
Therapeutic administration of LGG attenuates colonic tumor burden. (A) Graphic representation of the experimental outline for the induction of colonic tumors in C57BL/6 mice. Seven- to 8-week-old male mice were injected with AOM before being subjected to 3 rounds of 1-week exposure to 2% DSS and a 2-week recovery period. After 3 cycles of DSS, and between weeks 8 and 10 of the experiment, tumors begin to form within the colonic epithelium of treated mice. At week 10, groups of mice were supplemented with HBSS, BC, or LGG, or by oral gavage daily for a total of 6 weeks. Mice were killed at week 16. (B) Incidence of rectal bleeding or rectal prolapse between weeks 10 and 16 (the treatment period) of mice described in panel A. (C) Quantification of tumor burden at death at week 16 after removal of colon and opening the colon longitudinally with visual examination of the colon of groups of mice described in panel A. (D) Representative images of colons bearing colonic tumor with red arrows denoting areas with tumors as evaluated by a pathologist. (E) H&E sections of Swiss-rolled colonic tissue at 16 weeks of mice described in panel A, with representative images of entire colon (upper panels) and high-powered view (lower panels) of the area shown in the rectangle of upper panels. (F) Quantification of dysplastic epithelium at 16 weeks of mice described in panel A. (G) Immunofluorescence analysis for the detection of CD8 T cell (green) DNA (blue) within colonic polyps at 16 weeks of mice described in panel A. (H) Quantification of CD8 T cells (green) within colonic polyps at 16 weeks of mice described in panel F. One-way analysis of variance and Martel–Cox used for statistics and represented as follows: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. n = 14, 7, and 14 for HBSS, BC, and LGG, respectively. DAPI, 4′,6-diamidino-2-phenylindole, HPF, high-power field; IP, intraperitoneally.
LGG Requires CD8 T Cells to Elicit Reduction of Colonic Tumor Burden
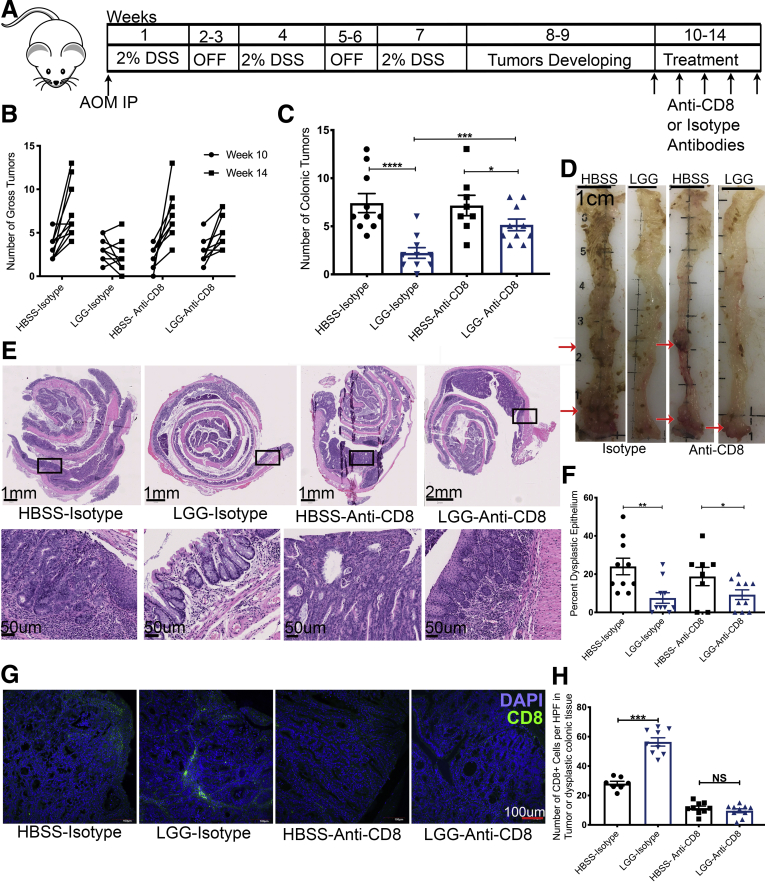
Previous studies have reported that LGG uses epithelial-specific mechanisms for reduced tumor burden when administered preventatively. However, because of our observation of higher infiltrating CD8 T cells, we tested whether LGG requires CD8 T cells for its attenuation of colonic tumors when administered therapeutically. To this end, we again performed a DSS-AOM experiment, but in conjunction with bacterial administration, we intraperitoneally injected anti-CD8 antibodies or isotype control antibodies to deplete CD8 T cells (Figure 6A). First, we verified that an intraperitoneal injection of anti-CD8 antibodies would be able to deplete CD8 T cells in the colon and mLn (data not shown). In addition, to mitigate variability in tumor burden induced by DSS-AOM between groups, we performed colonic endoscopy on all mice before treatment to establish baseline tumor burden. Furthermore, LGG treatment was shortened to 4 weeks because humoral responses against intraperitoneally injected antibodies may be detected after 4 weeks. At death, we observed that LGG significantly reduces tumor burden as compared with HBSS controls in isotype control-treated mice. However, this effect was reduced significantly when CD8 T cells were depleted (Figure 6B–D). H&E staining and histologic analysis corroborated the results and detected fewer dysplastic epithelium when LGG was administered with isotype controls (Figure 6E and F). Likewise, immunofluorescent analysis for CD8 T cells confirmed our findings that LGG increases intratumoral or intradysplastic epithelial CD8 T cells in mice treated with isotype controls and confirmed the depletion of CD8 T cells within tumors in anti-CD8 antibody groups (Figure 6G and H). Interestingly, LGG was able to reduce the tumor burden somewhat, even when CD8 T cells were depleted, but to a significantly lesser extent compared with LGG with CD8 T cells present. This lesser response in the absence of CD8 T cells may be owing to reported influences of LGG on tumor burden via epithelial-mediated mechanisms.29,31,33 In addition, the speculated epithelial responses are corroborated by the observation that mice administered LGG and anti-CD8 had a similar amount of dysplastic epithelium as LGG with CD8 T cells present, suggesting that some part of the tumor reduction and the effect on dysplastic epithelium may be owing in part to LGG’s effect on the epithelium (Figure 6C and F). Together, these data suggest that the novel immune mechanism initiated by LGG described in this article plays a dominant role in reducing colonic tumor burden when administered therapeutically.
Figure 6.
LGG requires CD8 T cells to elicit a strong reduction of colonic tumors. (A) Graphic representation of the experimental outline for the induction of colonic tumors in C57BL/6 mice. Seven- to 8-week-old mice were injected with AOM before being subjected to 3 rounds of 1-week exposure to 2% DSS and a 2-week recovery period. After 3 cycles of DSS, and between weeks 8 and 10 of the experiment, tumors begin to form within the colonic epithelium of treated mice. Tumors were quantified by miniature colonoscope at week 10, before the start of treatment period at week 10, in which groups of mice were supplemented with HBSS, BC, or LGG, or by oral gavage daily for a total of 4 weeks period. Tumors were quantified further by miniature colonoscope at week 14, at the end of the 4-week treatment period. In addition, during the 4-week treatment period, groups of mice were administered either an anti-CD8 or an isotype control weekly for 4 weeks. Mice were killed at week 14. (B) Numeration of tumor burden as detected by mature colonoscopy at weeks 10 and 14 of mice described in panel A. (C) Quantification of tumor burden at death at week 14 after removal of colon and opening the colon longitudinally with visual examination of the colons of groups of mice described in panel A. (D) Representative images of colons bearing colonic tumor with red arrows denoting areas with tumors as evaluated by a pathologist. (E) H&E sections of Swiss-rolled colonic tissue at 14 weeks of mice described in panel A, with representative images of entire colon (upper panels) and high-powered view (lower panels) of the area shown in the rectangle of upper panels. (F) Quantification of dysplastic epithelium at 14 weeks of mice described in panel A. (G) Immunofluorescence analysis for the detection of CD8 T cell (green) DNA (blue) within colonic polyps at 14 weeks of mice described in panel A. (H) Quantification of CD8 T cells (green) within colonic polyps at 14 weeks of mice described in panel F. One-way analysis of variance used for statistics and represented as follows: ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. n = 9, 9, 8, and 10 for HBSS-isotype, LGG-isotype, HBSS-anti-CD8, and LGG-anti-CD8, respectively. DAPI, 4′,6-diamidino-2-phenylindole.
Discussion
An increasing body of scientific literature reports on the role of the microbiome in cancer development, in cancer therapy, and in augmenting currently approved treatment modalities. In particular, a trio of articles have reported that the gut microbiome influences the efficacy of anti–pPD-1 in metastatic melanoma patients.14, 15, 16 They reported that contrasting commensal microbial communities exist within anti–PD-1–responding patients and anti–PD-1–nonresponding patients. Data from these reports have shown an absence of lactobacilli in anti–PD-1–nonresponding patients, whereas lactobacilli was present in responding patients. Furthermore, a subsequent article reported that a defined set of commensal microorganisms isolated from human feces were capable of inducing CD8 T cells in the intestine.34 These reports established the principle that members of the commensal microbiota can elicit immunomodulatory effects for the benefit of patients undergoing cancer treatment. Based on these published data as scientific premise, we reasoned that dietary supplementation with a human commensal probiotic that harbors the capacity to elicit expansion of intestinal CD8 T cells may be efficacious at inhibiting tumor growth.
We reported a novel mechanism whereby a constituent of the human gut microbiome and a widely used probiotic, LGG, activated a CD8 T-cell response. We showed that administration of LGG to mice expanded the number of total and effector CD8 T cells in a response localized to the intestine. Indeed, LGG also can expand CD4 T cells, showing that both cell types are influenced by LGG. Moreover, the expansion of CD4 T cells may function in promoting the efficacy of the CD8 T-cell response because we saw increased IL12 by antibody array and reverse-transcription polymerase chain reaction (RT-PCR). IL12 is known to regulate granzyme-B expression, which we observed was increased in CD8 T cells from LGG-treated animals. This expansion was dependent on DCs, and, more specifically, required TLR2 expression on DCs to expand T cells. In contrast to previous reports that treated LGG prophylactically,29,30,33 we showed that LGG given therapeutically in a mouse model of colonic tumors can confer an antitumor response. To show that LGG-induced expansion of CD8 T cells was required for the anti-tumor response, we depleted CD8 T cells in conjunction with LGG administration and observed an attenuation of LGG’s antitumor effect. These data support the notion that supplementation with LGG holds the promise of augmenting cancer therapy when administered in combination with currently approved therapeutics.
Published literature has speculated that metabolites released by the microbiome may be the mechanisms whereby certain gut microbes elicit their effects on T cells.34 Indeed, our research group reported that LGG can modify the hepatic transcriptome and other distal host sites via the generation of gut microbe–derived metabolites.17,35 However, we show conclusive evidence that LGG-induced expansion of T cells requires TLR2 expression by DCs. Indeed, a recent report showed that enhanced immunotherapy by the microbiome requires TLR2 and may be the result of the same mechanism described in this study.36 Moreover, in heathy mice and in mice subjected to the DSS-AOM and MSH2 models, LGG increased the CD8 T-cell response by approximately 2-fold, with no detectable negative pathologic damage to the normal colonic epithelium. Importantly, this effect was preserved when heat-killed LGG was administered, indicating that the metabolic activity of LGG was not required for this response. In addition, in vitro studies in which BMDCs were exposed to pure cultures of LGG detected TLR2-dependent activation of DCs. These data point to the conclusion that the sentinel action of TLR2 allows DCs to sense a heat-resistant LGG microbe-associated molecular pattern, and that this is the most likely stimulatory mechanism of DCs in our studies. Therefore, we speculate that under healthy conditions, LGG-activated DCs likely will sample LGG-derived proteins and products, as well as other bacterial proteins and products generated by the extant microbiome and the healthy colonic epithelium. Notably, despite the ensuing increase of CD8 T cells that we detected, no pathologic tissue damage was observed, possibly because of the inhibitory mechanisms against normal host factors. Indeed, we did detect a nonsignificant increase in IL10 by antibody array, which may aid in preventing immunopathologic damage in healthy mice. Alternatively, when tumors are present, LGG-activated DCs also will sample mutated host proteins, and the responding CD8 T cells will initiate an antitumor response. In this situation, we do not discount the possibility that LGG and the extant microbiome may synergize and augment T-cell activation and the antitumor response. Nevertheless, we show that LGG induced a potent antitumorigenic response, without any negative effects during the DSS-AOM model and during homeostatic conditions. We propose that these data serve as preclinical evidence for the rational use of LGG in relevant clinical situations.
The data reported here contrast with some published reports on immune response to lactobacilli, but importantly are consistent with several published data investigating the immunomodulatory activity of LGG.17, 18, 19, 20, 21, 22, 23, 37 Previous reports on the anti-inflammatory influences of LGG showed that LGG expanded regulatory T-cell populations in the gut and in the bone marrow.17 In that report, it was shown that LGG induced anti-inflammatory mechanisms via generation of the short-chain fatty acid butyrate. Interestingly, it was reported that LGG itself does not produce butyrate, but provides a substrate (lactic acid) to other members of the microbiome such as Clostridia species, which convert lactate to butyrate.17 Of note, in the current study, we administered LGG daily but did not detect any changes in the levels of IL10 or the number of regulatory T cells in our models. However, direct contact of LGG with immune cells in vitro induce proinflammatory responses, which illustrate that LGG can evoke proinflammatory responses in certain contexts. In our hands, observations of LGG’s immunomodulation consistently showed a proinflammatory context under both homeostatic conditions and colonic cancer, but these effects might change depending on the disease context. In addition, many reports illustrating that lactobacilli and LGG are protective in settings of chronic inflammation show that this protection is mediated through effects on the colonic epithelium.26,38,39 Therefore, LGG’s effects on the epithelium must be considered as well to fully understand all dynamics at play.
Because contrasting commensal microbial communities exist within anti–PD-1–responding patients and anti–PD-1–nonresponding patients, and because responding patients have a more robust microbiome-induced CD8 T-cell response,15 it is practical to speculate about the extent to which further supplementation with LGG or any other CD8 T-cell–inducing bacteria may enhance immunotherapy efficacy and breadth. Because of heterogeneity in microbiome community structures across cultural and ethnic populations, using 16S ribosomal DNA-based methodology to identify a human microbiome community structure that induces an increased basal CD8 T-cell response in all individuals may be challenging. This would be confounded further, if, as was reported previously, a consortium of bacteria is necessary to expand CD8 T-cell numbers in the intestine.34 Until keystone bacterial species that expand CD8 T-cell numbers in the intestine are identified and are faithfully detectable, the lack of clearly defined microbiome that enhances CD8 T-cell responses will continue to be a barrier to progress in this field. Nevertheless, because dietary supplementation with LGG is relatively cheap and easy to administer, it may be asked if it would be recommended that all patients undergoing immunotherapy for colonic cancer should be supplemented with LGG or a similar CD8 T-cell–enhancing bacterium or consortium of bacteria. Indeed, a report showed that Lactobacillus acidophilus lysates can synergize with anti-Cytotoxic T-lymphocyte associated protein 4 (CTLA4) immunotherapy in mice if administered immediately after colitis.40 This could be considered for nonresponding immunotherapy patients, especially because there appears to be little to no negative effects of LGG supplementation in our mouse models of tumorigenesis, as well as no evidence of increased bacteremia by lactobacilli over the past few decades despite the increasingly widespread use of probiotic supplements that include lactobacilli.41
Together, we show data to support the use of bacteria-based modalities to treat CRC. This approach of using probiotics was reported previously to limit tumor burden only when the beneficial bacteria was administered prophylactically.29, 30, 31, 32, 33 We show data that therapeutic administration of LGG limits tumor burden, thereby offering a potential interventional therapy for CRC in conjunction with other modalities. Before the use of beneficial commensal bacteria in clinical trials, it is necessary to fully understand the host mechanisms at play. Our identification of the mechanism of how LGG elicits its efficacious effects on CRC adds to the growing body of scientific evidence for utilization of probiotics in cancer treatment and directly may inform decisions related to dosing and timing for the use of LGG in CRC clinical trials.
Methods
Animals
C57BL/6 mice (stock #000664) were obtained from Jackson Laboratories (Farmington, CT) and maintained in Emory’s Whitehead Vivarium or Emory’s Gnotobiotic Animal Core located in Emory’s Health Science Research Building Vivarium (Atlanta, GA). BATF3 knockout mice (stock #013755), TLR2 knockout mice (stock #004650), Villin Cre mice (stock #004586), and MSH2loxP mice (stock #016231) also were obtained from Jackson Laboratories. All experiments were performed with the approval of the Institutional Animal Care and Use Committee.
Bacterial Cultures and Growth Conditions
LGG (ATCC 53103) was grown in de Man, Rogosa, and Sharpe (MRS) broth at 37°C without shaking for 16 hours before administration. BC (laboratory strain) was grown in brain heart infusion media with shaking at 37°C for 16 hours before administration.35 Bacteria were centrifuged at 3000g for 5 minutes, the supernatant was aspirated, and the pellet was washed with one volume HBSS. This was repeated for a total of 3 washes. The bacteria then were resuspended to a final concentration of 2 × 109 colony-forming units/mL.
Administration of Bacteria
Mice were gavaged with commensals as described previously.42 Briefly, LGG, BC, or vehicle control, HBSS, were orally gavaged to mice every day in the midafternoon at a dose of 2 × 108 colony-forming units at a volume of 100 uL, with control mice receiving 100 uL of HBSS.
CFSE-Labeling of Bacteria
Bacteria were grown as mentioned earlier. However, after the third wash, bacteria were resuspended in 10 mL of Cell-Trace CFSE (ThermoFisher, Waltham, MA) and diluted 1:1000 in HBSS per the manufacturer’s instructions (heat-killed LGG was heat-killed before CFSE staining). Bacteria in CFSE solution were incubated at 37°C in the dark for 20 minutes and then centrifuged at 3000g for 5 minutes at room temperature to pellet, which visually will be yellow. Bacteria were washed 3 times with HBSS to remove any unbound dye. In addition, to control for any unbound dye, an empty 15-mL tube underwent the CFSE protocol and was administered to the HBSS group. Fluorescence was checked by a plate reader or FCM to verify fluorescence at 488 nm.
Isolation of Colonic Lamina Propria Lymphocytes
We performed protocols as previously described.43 Briefly, colons were extracted by cutting between the proximal colon and the cecum and between the distal colon and the anal verge. Colons were teased apart from the mesentery and flushed of luminal contents. Normally, colons were opened longitudinally and then cut in half longitudinally, with half going to isolation of colonic lamina propria lymphocytes and the other half for immunofluorescence. The colon was weighed, cut into small 1-cm pieces, and placed into a 50-mL conical tube with epithelial digestion buffer consisting of 1× HBSS with 5% fetal bovine serum (FBS), 5 mmol/L EDTA, and 10 mmol/L HEPES. Tubes were placed into a shaker at 200 rpm for 20 minutes at 37°C. A wire-mesh strainer was used to separate pieces of colon from buffer with a collection tube collecting epithelial cells and intraepithelial lymphocytes. This step was repeated for a total of 40 minutes of digestion. After the second straining, colonic pieces were washed briefly with RPMI-1640 media. The pieces then were minced with a clean razorblade and placed into a new 50-mL conical tube with lamina propria digestion buffer consisting of RPMI-1640 media with 10% FBS, 1 mg/mL collagenase type IV, and 50 ug/mL DNase I. Tubes were placed into a shaker at 200 rpm for 15 minutes at 37°C. Afterward, contents were emptied into a 70-μm cell strainer and contents were pushed through with a syringe stopper. Cells then were pelleted by centrifugation at 300g for 5 minutes at 4°C and resuspended in 1 mL complete media. Immune cells were isolated from total cell populations by a Percoll gradient: 90% Percoll overlaid with 30% Percoll. Cells were placed on top and centrifuged at 670g for 30 minutes at room temperature with acceleration and brakes set to slow. Immune cells are found as a band at the 90:30 interface, with remaining cells such as fat at the top. The cells at the top were aspirated, while leaving immune cells undisturbed. Fresh media was added to dilute the Percoll gradient and spun down at 300g for 5 minutes at 4°C. Pelleted cells were resuspended in media and pushed through a 40-μm filter and either immediately analyzed by flow cytometry or frozen in complete media with 10% dimethyl sulfoxide.
Antibody Array
C57Bl/6 mice were obtained from Jackson Laboratories and maintained in specific-pathogen free conditions inside Emory’s Gnotobiotic Animal Core (Atlanta, GA). Mice were orally gavaged every day with LGG, BC, or HBSS for 2 weeks, and kept inside an ISOcageP Bioexclusion system (Techniplast, West Chester, PA) maintained in a Biosafetly level 2 (BSL-2) facility. Upon killing, flash-frozen whole colons were given to RayBiotech, Inc (Peachtree Corners, GA) for protein extraction and cytokine analysis using the Quantibody Mouse Cytokine Array Q4000 (Raybiotech).
BMDC Generation and Bacterial Incubation
BMDCs were generated and activated as previously described.21,44 In short, the femur and tibia were removed from either WT or TLR2-/- mice with the bone marrow being removed in a sterile hood. Red blood cells were lysed and 5 × 105 progenitor cells/mL were resuspended and plated in RPMI 1640 supplemented with 10% FBS and 10 ng/mL Granulocyte-Macrophage Colony Stimulating Factor GM-CSF (Biolegend, San Diego, CA). BMDCs were cultured for 6 days at 37°C in 5% CO2 with replenishing of complete media and cytokines on days 2 and 4. On day 6, BMDCs were removed and replated at 5 × 105 cells/well. After 2 hours, BMDCs were incubated with a concentration of 10:1 BC, LGG, heat-killed LGG, or HBSS for 4 hours. At 4 hours, 200 ug/mL gentamycin was added to kill all extracellular bacteria and cells were washed 3 times with HBSS. Fresh media then was given and BMDCs were incubated for an additional 20 hours. BMDCs then were stained and analyzed by FCM for expression of CD80 and CD86.
Flow Cytometry
Flow cytometry was performed as previously described.43 Briefly, a single-cell suspension of colon, lymph node, or spleen was placed in a round-bottom 96-well plate and centrifuged at 300g for 5 minutes at 4°C. Supernatant was flicked off and cells were resuspended in 100 uL antibody master mix. Antibody master mix contained desired antibodies run on 2 different panels (the first panel consisted of Allophycocyanin(APC)-CD3, Peridinin Chlorophyll Protein Complex(PerCP)-CD4, Brilliant Violet (BV)-605-CD8, Phycoerythrin(PE)-CD44, BV650-CD62L, BV510-CD25, B421-CXCR3, PE-Texas-Red(TR)-CD28, and fluorescein isothiocyanate (FITC)–Granzyme-B; the second panel consisted of BV421-CD11b, PE-CD11c, Alexa700-F4/80, PerCP-Major Histocompaibiltiy Complex II(MHCII), BV650-Ly6G, APC-CD19, PE-TR-CD80, BV510-CD86, and BV711-CD103; all from Biolegend, San Diego, CA) at 1:100 dilution and the LIVE/DEAD Fixable near-Infrared-Inf I staining kit at 1:1000 dilution (ThermoFischer) in fluorescence-activated cell sorter (FACS) buffer containing 1× phosphate-buffered saline (PBS) with 5% FBS and 0.5 mmol/L EDTA. Cells were stained for 1 hour at 4°C in the dark. Afterward, cells were centrifuged and washed with FACS buffer for a total of 3 times. In addition, for granzyme B staining, we used the True-Nuclear Transcription Factor Buffer Set (Biolegend) according to the manufacturer’s instructions. After staining, cells were resuspended in a final concentration of 200 uL FACS buffer and analyzed by a LSRII Flow Cytometer (BDbiosciences, Franklin Lakes, NJ). Data were analyzed using FlowJo software (Flow Jo LLC, Ashland, OR). The gating strategy for T cells consisted of cells/single cells/live cells/CD3+, and then analyzed for CD4 and CD8 with subsequent effector status and other molecules. The gating strategy for DCs consisted of cells/single cells/live cells/CD3-CD19-/MHC II+/CD11c+, and then analyzed for activation or for CFSE uptake. Cell numbers were calculated using Precision Count Beads (Biolegend). Compensation was performed via single-color controls using UltraComp eBeads Compensation Beads (ThermoFischer) and for CFSE compensation was performed with stained and unstained bacteria, with compensation matrices created in FlowJo software.
Immunofluorescence
Colon samples for immunofluorescence were Swiss-rolled and fixed in 10% formalin and then processed and embedded in paraffin. Five-micron sections were cut and underwent a deparaffination process followed by a citrate antigen retrieval step before being permeabilized in 0.5% Triton X-100 (ThermoFisher, Waltham, MA) in PBS and blocked with 5% normal goat serum. The CD8 monoclonal rat antibody (eBioscience, San Diego, CA) was used at 1:100 in PBS with 0.1% Triton-X and 5% normal goat serum. Samples were incubated in primary antibody overnight at 4°C. Samples then were washed 3 times and incubated for 1 hour in fluorochrome-conjugated secondary antibody. Samples were again washed and incubated with 4′,6-diamidino-2-phenylindole at a concentration of 1:10,000 in PBS for 5 minutes. Samples were mounted using Prolong diamond antifade (ThermoFisher, Waltham, MA) and imaged on an Olympus FV1000 confocal microscope (Olympus, Center Valley, PA) at 20×. At least 3 sections and 3 different images were taken for each sample.
Histology
Colon samples were Swiss-rolled upon death and fixed in 10% formalin and then processed and embedded in paraffin. Five-micron sections were cut and underwent H&E staining as performed by Emory University’s Cancer Tissue and Pathology core (Atlanta, GA). Stained slides were examined by a board-certified pathologist for dysplasia and the percentage of the dysplastic epithelium of the section was calculated by visual inspection.
Transcriptional Analysis
For transcriptional analysis, 1 cm of the proximal colon was dissected and snap-frozen in liquid nitrogen. Colon then was homogenized by mechanical disruption using a MagnaLyser (Roche, Basel, Switzerland) with ceramic MagnaLyser beads (Roche) with 1 mL TRIzol (ThermoFisher, Waltham, MA). RNA was prepared according to the TRIzol manufacturer’s instructions. Complementary DNA was synthesized using the iScript Complementary DNA synthesis kit (BioRad) and the manufacturer’s instructions were performed using 1 ug RNA. Reverse-transcription PCR was performed using SybrGreen supermix (BioRad, Hercules, CA), with a 2-step amplification protocol while using the following primers: CXCL9: forward: CGAGGCACGATCCACTACAA; reverse: CCGGATCTAGGCAGGTTTGA; CXCL10: forward: TGCGTGGCTTCACTCCAGTT; reverse: TCCTGCCCACGTGTTGAGAT; IL12p40: forward: TCTTCAAAGGCTTCATCTGCAA; reverse: ACAGCACCAGCTTCTTCATCA; and TLR2: forward: GGGCTTCACTTCTCTGCTTT; reverse: TCCTCTGAGATTTGACGCTTTG. Data were analyzed using the delta, delta cycle threshold D method.
Intestinal MSH2 Cancer Model
For intiating colonic tumors, we utilized the well-characterized model for genetically-induced colonic tumors known as the MSH2 colonic cancer model.28 Briefly, intestinal-specific depletion of exon 12 of MSH2 was generated by crossing MSH2loxP with Villin Cre mice until homozygosity. Genotypes were confirmed by PCR (Transnetyx, Cordova, TN) and mice then were aged to approximately 7 months. At this point, mice began receiving either HBSS, LGG, or BC daily by oral gavage for 6 weeks. At death, colons were removed, opened longitudinally, and examined for colonic tumors. Examination of tissue was performed with training from a board-certified pathologist.
DSS-AOM Model
We performed the well-characterized chemical model of DSS-AOM.45 Briefly, we intraperitoneally injected AOM at a dose of 60 mg/kg from a solution of 10 mg/mL, which equated to roughly 100–150 uL AOM for mice ranging from 20–25 g. Mice were 7–8 weeks old at the time of injection. Three days after injection of AOM, drinking water was replaced with 2% DSS for 7 days. After induction of colitis, mice were allowed to recover for 2 weeks on normal drinking water. Colitis was induced by DSS for a total of 3 times. After the third round of colitis, mice were allowed to recover for 2 weeks on normal drinking water before undergoing endoscopy to verify tumor burden. At this point, intervention with either monoclonal antibodies or bacteria was introduced for 4–6 weeks, depending on the experiment. At death, colons were removed, opened longitudinally, and examined for colonic tumors. Examination of tissue was performed with training from a board-certified pathologist.
Endoscope
All procedures and postanesthesia care were performed in accordance within Institutional Animal Care and Use Committee guidelines. Each mouse was anesthetized with 3% isoflurane and maintained with 1.5% isoflurane in an oxygen/air mixture by using a gas anesthesia mask. Body temperature was maintained during the procedure at 37°C with a homeothermic blanket (Harvard Apparatus, Holliston, MA). For monitoring tumor burden, a high-resolution mouse video endoscopic system was used (Karl Storz Endoskope, Tuttlingen, Germany). The system consists of a miniature endoscope (outer diameter, 1.9 mm), a xenon light source, a triple-chip camera, and an air pump for regulated inflation of the mouse colon (Karl Storz). The endoscopic procedure was viewed on a color monitor and digitally recorded for postprocedure review with recording of the ileocecal valve all the way to the anus of each mouse. Videos were reviewed by a board-certified pathologist and tumor burden was established by their identification of individual tumors in the videos of each mouse.
Depletion of CD8 T Cells
CD8 T cells were depleted by intraperitoneal injection of CD8a monoclonal antibody (clone: YTS 169.4; BioXCell, Lebanon, NH). A total of 200 ug monoclonal antibody CD8a or isotype control (rat IgG2b; clone: LTF-2; BioXCell) was given 2 days before administration of bacteria and was administered every 6–7 days afterward.
Quantification and Statistics
Graphs are shown as means ± SEM. Statistical analyses were performed using GraphPad Prism software. For comparisons of 2 groups, the unpaired Student t test was used. For comparisons of groups of 3 or more, 1-way analysis of variance was used, followed by the Dunnett multiple comparison test. For incidence of rectal prolapse, the Martel–Cox test was used. Statistical parameters are stated within the figure legends.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
CRediT Authorship Contributions
Joshua A Owens, BSc (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Bejan J Saeedi, PhD (Investigation: Supporting)
Crystal R Naudin, PhD (Investigation: Supporting; Methodology: Supporting)
Sarah Hunter-Chang, BSc (Investigation: Supporting)
Maria E Barbian, MD (Investigation: Supporting)
Richard U Eboka, BSc (Investigation: Supporting)
Lauren Askew, BSc (Investigation: Supporting)
Trevor M Darby, PhD (Investigation: Supporting)
Brian S Robinson, MD PhD (Investigation: Supporting)
Rheinallt M Jones, PhD (Conceptualization: Equal; Funding acquisition: Lead; Project administration: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported in part by National Institutes of Health grants R01DK098391 and R01CA179424 (R.M.J.), F31CA247415 (J.A.O.), and F30DK117570 (B.J.S.). Also supported by American Heart Association Fellowship19POST34370006 (C.R.N.), the Marshall Klaus Perinatal Research Award and Emory Pediatrics Warshaw Fellow Award (M.E.B.), the American Gastroenterological Association (T.M.D.), and by Research Training in Translational Gastroenterology and Hepatology training grant T32DK108735 (B.S.R.).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012: global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 4.Overman M.J., Lonardi S., Wong K.Y.M., Lenz H.-J., Gelsomino F., Aglietta M., Morse M.A., Van Cutsem E., McDermott R., Hill A., Sawyer M.B., Hendlisz A., Neyns B., Svrcek M., Moss R.A., Ledeine J.-M., Cao Z.A., Kamble S., Kopetz S., André T. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 5.Overman M.J., Ernstoff M.S., Morse M.A. Where we stand with immunotherapy in colorectal cancer: deficient mismatch repair, proficient mismatch repair, and toxicity management. Am Soc Clin Oncol. 2018;38:239–247. doi: 10.1200/EDBK_200821. [DOI] [PubMed] [Google Scholar]
- 6.Brown S.D., Warren R.L., Gibb E.A., Martin S.D., Spinelli J.J., Nelson B.H., Holt R.A. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lennerz V., Fatho M., Gentilini C., Frye R.A., Lifke A., Ferel D., Wölfel C., Huber C., Wölfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 9.Roxburgh C.S.D., McMillan D.C. The role of the in situ local inflammatory response in predicting recurrence and survival in patients with primary operable colorectal cancer. Cancer Treat Rev. 2012;38:451–466. doi: 10.1016/j.ctrv.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Gryfe R., Kim H., Hsieh E.T., Aronson M.D., Holowaty E.J., Bull S.B., Redston M., Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 11.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P.-H., Trajanoski Z., Fridman W.-H., Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 12.Naito Y., Saito K., Shiiba K., Ohuchi A., Saigenji K., Nagura H., Ohtani H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 13.Jansen C.S., Prokhnevska N., Master V.A., Sanda M.G., Carlisle J.W., Bilen M.A., Cardenas M., Wilkinson S., Lake R., Sowalsky A.G., Valanparambil R.M., Hudson W.H., McGuire D., Melnick K., Khan A.I., Kim K., Chang Y.M., Kim A., Filson C.P., Alemozaffar M., Osunkoya A.O., Mullane P., Ellis C., Akondy R., Im S.J., Kamphorst A.O., Reyes A., Liu Y., Kissick H. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 2019;576:465–470. doi: 10.1038/s41586-019-1836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., Cogdill A.P., Zhao L., Hudgens C.W., Hutchinson D.S., Manzo T., Petaccia de Macedo M., Cotechini T., Kumar T., Chen W.S., Reddy S.M., Szczepaniak Sloane R., Galloway-Pena J., Jiang H., Chen P.L., Shpall E.J., Rezvani K., Alousi A.M., Chemaly R.F., Shelburne S., Vence L.M., Okhuysen P.C., Jensen V.B., Swennes A.G., McAllister F., Marcelo Riquelme Sanchez E., Zhang Y., Le Chatelier E., Zitvogel L., Pons N., Austin-Breneman J.L., Haydu L.E., Burton E.M., Gardner J.M., Sirmans E., Hu J., Lazar A.J., Tsujikawa T., Diab A., Tawbi H., Glitza I.C., Hwu W.J., Patel S.P., Woodman S.E., Amaria R.N., Davies M.A., Gershenwald J.E., Hwu P., Lee J.E., Zhang J., Coussens L.M., Cooper Z.A., Futreal P.A., Daniel C.R., Ajami N.J., Petrosino J.F., Tetzlaff M.T., Sharma P., Allison J.P., Jenq R.R., Wargo J.A. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., Fidelle M., Flament C., Poirier-Colame V., Opolon P., Klein C., Iribarren K., Mondragón L., Jacquelot N., Qu B., Ferrere G., Clémenson C., Mezquita L., Masip J.R., Naltet C., Brosseau S., Kaderbhai C., Richard C., Rizvi H., Levenez F., Galleron N., Quinquis B., Pons N., Ryffel B., Minard-Colin V., Gonin P., Soria J.-C., Deutsch E., Loriot Y., Ghiringhelli F., Zalcman G., Goldwasser F., Escudier B., Hellmann M.D., Eggermont A., Raoult D., Albiges L., Kroemer G., Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi A.M., Yu M., Darby T.M., Vaccaro C., Li J.-Y., Owens J.A., Hsu E., Adams J., Weitzmann M.N., Jones R.M., Pacifici R. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018;49:1116–1131.e7. doi: 10.1016/j.immuni.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia L., Wu R., Han N., Fu J., Luo Z., Guo L., Su Y., Du J., Liu Y. Porphyromonas gingivalis and Lactobacillus rhamnosus GG regulate the Th17/Treg balance in colitis via TLR4 and TLR2. Clin Transl Immunol. 2020;9:e1213. doi: 10.1002/cti2.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirpuri J., Sotnikov I., Myers L., Denning T.L., Yarovinsky F., Parkos C.A., Denning P.W., Louis N.A. Lactobacillus rhamnosus (LGG) regulates IL-10 signaling in the developing murine colon through upregulation of the IL-10R2 receptor subunit. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harata G., He F., Hiruta N., Kawase M., Kubota A., Hiramatsu M., Yausi H. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett Appl Microbiol. 2010;50:597–602. doi: 10.1111/j.1472-765X.2010.02844.x. [DOI] [PubMed] [Google Scholar]
- 21.Cai S., Kandasamy M., Rahmat J.N., Tham S.M., Bay B.H., Lee Y.K., Mahendran R. Lactobacillus rhamnosus GG activation of dendritic cells and neutrophils depends on the dose and time of exposure. J Immunol Res. 2016;2016 doi: 10.1155/2016/7402760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., Wu Y., Liu R., Xu H., Mei X., Shang Q., Liu S., Yu D., Li W. Lactobacillus rhamnosus GG promotes M1 polarization in murine bone marrow-derived macrophages by activating TLR2/MyD88/MAPK signaling pathway. Anim Sci J. 2020;91:e13439. doi: 10.1111/asj.13439. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Liu L., Moore D.J., Shen X., Peek R.M., Acra S.A., Li H., Ren X., Polk D.B., Yan F. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 2017;10:373–384. doi: 10.1038/mi.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riehl T.E., Alvarado D., Ee X., Zuckerman A., Foster L., Kapoor V., Thotala D., Ciorba M.A., Stenson W.F. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut. 2019;68:1003–1013. doi: 10.1136/gutjnl-2018-316226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Zhou Z., Li Y., Zhou L., Ding Q., Xu L. Isolated exopolysaccharides from Lactobacillus rhamnosus GG alleviated adipogenesis mediated by TLR2 in mice. Sci Rep. 2016;6:36083. doi: 10.1038/srep36083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryu S.-H., Park J.-H., Choi S.-Y., Jeon H.-Y., Park J.-I., Kim J.-Y., Ham S.-H., Choi Y.-K. The probiotic Lactobacillus prevents Citrobacter rodentium-induced murine colitis in a TLR2-dependent manner. J Microbiol Biotechnol. 2016;26:1333–1340. doi: 10.4014/jmb.1602.02004. [DOI] [PubMed] [Google Scholar]
- 27.de Miranda N.F.C.C., Goudkade D., Jordanova E.S., Tops C.M.J., Hes F.J., Vasen H.F.A., van Wezel T., Morreau H. Infiltration of Lynch colorectal cancers by activated immune cells associates with early staging of the primary tumor and absence of lymph node metastases. Clin Cancer Res. 2012;18:1237–1245. doi: 10.1158/1078-0432.CCR-11-1997. [DOI] [PubMed] [Google Scholar]
- 28.Kucherlapati M.H., Lee K., Nguyen A.A., Clark A.B., Hou H., Rosulek A., Li H., Yang K., Fan K., Lipkin M., Bronson R.T., Jelicks L., Kunkel T.A., Kucherlapati R., Edelmann W. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 2010;138:993–1002.e1. doi: 10.1053/j.gastro.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Femia A.P., Luceri C., Dolara P., Giannini A., Biggeri A., Salvadori M., Clune Y., Collins K.J., Paglierani M., Caderni G. Antitumorigenic activity of the prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis on azoxymethane-induced colon carcinogenesis in rats. Carcinogenesis. 2002;23:1953–1960. doi: 10.1093/carcin/23.11.1953. [DOI] [PubMed] [Google Scholar]
- 30.Goldin B.R., Gualtieri L.J., Moore R.P. The effect of Lactobacillus GG on the initiation and promotion of DMH-induced intestinal tumors in the rat. Nutr Cancer. 1996;25:197–204. doi: 10.1080/01635589609514442. [DOI] [PubMed] [Google Scholar]
- 31.Gamallat Y., Meyiah A., Kuugbee E.D., Hago A.M., Chiwala G., Awadasseid A., Bamba D., Zhang X., Shang X., Luo F., Xin Y. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed Pharmacother. 2016;83:536–541. doi: 10.1016/j.biopha.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Ni Y., Wong V.H.Y., Tai W.C.S., Li J., Wong W.Y., Lee M.M.L., Fong F.L.Y., El-Nezami H., Panagiotou G. A metagenomic study of the preventive effect of Lactobacillus rhamnosus GG on intestinal polyp formation in ApcMin/+ mice. J Appl Microbiol. 2017;122:770–784. doi: 10.1111/jam.13386. [DOI] [PubMed] [Google Scholar]
- 33.Walia S., Kamal R., Dhawan D.K., Kanwar S.S. Chemoprevention by probiotics during 1,2-dimethylhydrazine-induced colon carcinogenesis in rats. Dig Dis Sci. 2018;63:900–909. doi: 10.1007/s10620-018-4949-z. [DOI] [PubMed] [Google Scholar]
- 34.Tanoue T., Morita S., Plichta D.R., Skelly A.N., Suda W., Sugiura Y., Narushima S., Vlamakis H., Motoo I., Sugita K., Shiota A., Takeshita K., Yasuma-Mitobe K., Riethmacher D., Kaisho T., Norman J.M., Mucida D., Suematsu M., Yaguchi T., Bucci V., Inoue T., Kawakami Y., Olle B., Roberts B., Hattori M., Xavier R.J., Atarashi K., Honda K. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 35.Saeedi B.J., Liu K.H., Owens J.A., Hunter-Chang S., Camacho M.C., Eboka R.U., Chandrasekharan B., Baker N.F., Darby T.M., Robinson B.S., Jones R.M., Jones D.P., Neish A.S. Gut-resident lactobacilli activate hepatic Nrf2 and protect against oxidative liver injury. Cell Metab. 2020;31:956–968.e5. doi: 10.1016/j.cmet.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S.-H., Cho S.-Y., Yoon Y., Park C., Sohn J., Jeong J.-J., Jeon B.-N., Jang M., An C., Lee S., Kim Y.Y., Kim G., Kim S., Kim Y., Lee G.B., Lee E.J., Kim S.G., Kim H.S., Kim Y., Kim H., Yang H.-S., Kim S., Kim S., Chung H., Moon M.H., Nam M.H., Kwon J.Y., Won S., Park J.-S., Weinstock G.M., Lee C., Yoon K.W., Park H. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- 37.Davidson L.E., Fiorino A.-M., Snydman D.R., Hibberd P.L. Lactobacillus GG as an immune adjuvant for live-attenuated influenza vaccine in healthy adults: a randomized double-blind placebo-controlled trial. Eur J Clin Nutr. 2011;65:501–507. doi: 10.1038/ejcn.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciorba M.A., Riehl T.E., Rao M.S., Moon C., Ee X., Nava G.M., Walker M.R., Marinshaw J.M., Stappenbeck T.S., Stenson W.F. Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR-2/cyclo-oxygenase-2-dependent manner. Gut. 2012;61:829–838. doi: 10.1136/gutjnl-2011-300367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones R.M., Desai C., Darby T.M., Luo L., Wolfarth A.A., Scharer C.D., Ardita C.S., Reedy A.R., Keebaugh E.S., Neish A.S. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 2015;12:1217–1225. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuo Q., Yu B., Zhou J., Zhang J., Zhang R., Xie J., Wang Q., Zhao S. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci Rep. 2019;9:20128. doi: 10.1038/s41598-019-56661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salminen M.K., Tynkkynen S., Rautelin H., Saxelin M., Vaara M., Ruutu P., Sarna S., Valtonen V., Järvinen A. Lactobacillus bacteremia during a rapid increase in probiotic use of Lactobacillus rhamnosus GG in Finland. Clin Infect Dis. 2002;35:1155–1160. doi: 10.1086/342912. [DOI] [PubMed] [Google Scholar]
- 42.Darby T.M., Owens J.A., Saeedi B.J., Luo L., Matthews J.D., Robinson B.S., Naudin C.R., Jones R.M. Lactococcus Lactis subsp. cremoris is an efficacious beneficial bacterium that limits tissue injury in the intestine. iScience. 2019;12:356–367. doi: 10.1016/j.isci.2019.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owens J.A., Jones R.M. Immune cell isolation from murine intestine for antibody array analysis. Methods Mol Biol. 2021;2237:247–256. doi: 10.1007/978-1-0716-1064-0_21. [DOI] [PubMed] [Google Scholar]
- 44.Henry C.J., Grayson J.M., Brzoza-Lewis K.L., Mitchell L.M., Westcott M.M., Cook A.S., Hiltbold E.M. The roles of IL-12 and IL-23 in CD8+ T cell-mediated immunity against Listeria monocytogenes: insights from a DC vaccination model. Cell Immunol. 2010;264:23–31. doi: 10.1016/j.cellimm.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popivanova B.K., Kitamura K., Wu Y., Kondo T., Kagaya T., Kaneko S., Oshima M., Fujii C., Mukaida N. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118:560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]